Huntington's disease age-of-onset linked to polyglutamine aggregation nucleation
- PMID: 12186976
- PMCID: PMC129363
- DOI: 10.1073/pnas.182276099
Huntington's disease age-of-onset linked to polyglutamine aggregation nucleation
Abstract
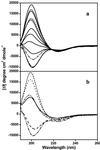
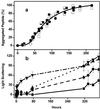
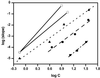
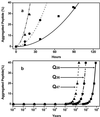
In Huntington's Disease and related expanded CAG repeat diseases, a polyglutamine [poly(Gln)] sequence containing 36 repeats in the corresponding disease protein is benign, whereas a sequence with only 2-3 additional glutamines is associated with disease risk. Above this threshold range, longer repeat lengths are associated with earlier ages-of-onset. To investigate the biophysical basis of these effects, we studied the in vitro aggregation kinetics of a series of poly(Gln) peptides. We find that poly(Gln) peptides in solution at 37 degrees C undergo a random coil to beta-sheet transition with kinetics superimposable on their aggregation kinetics, suggesting the absence of soluble, beta-sheet-rich intermediates in the aggregation process. Details of the time course of aggregate growth confirm that poly(Gln) aggregation occurs by nucleated growth polymerization. Surprisingly, however, and in contrast to conventional models of nucleated growth polymerization of proteins, we find that the aggregation nucleus is a monomer. That is, nucleation of poly(Gln) aggregation corresponds to an unfavorable protein folding reaction. Using parameters derived from the kinetic analysis, we estimate the difference in the free energy of nucleus formation between benign and pathological length poly(Gln)s to be less than 1 kcal/mol. We also use the kinetic parameters to calculate predicted aggregation curves for very low concentrations of poly(Gln) that might obtain in the cell. The repeat-length-dependent differences in predicted aggregation lag times are in the same range as the length-dependent age-of-onset differences in Huntington's disease, suggesting that the biophysics of poly(Gln) aggregation nucleation may play a major role in determining disease onset.
Figures

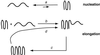
References
-
- Cummings C J, Zoghbi H Y. Hum Mol Genet. 2000;9:909–916. - PubMed
-
- Myers R H, Marans K S, MacDonald M E. In: Genetic Instabilities and Hereditary Neurological Diseases. Wells R D, Warren S T, editors. San Diego: Academic; 1998. pp. 301–323.
-
- DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. - PubMed
-
- Reddy P H, Williams M, Tagle D A. Trends Neurosci. 1999;22:248–255. - PubMed
-
- Wetzel R, Chen S, Berthelier V, Yang W. Soc Neurosci Abstr. 2001;27:578.12.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

