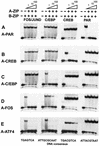
Classification of human B-ZIP proteins based on dimerization properties
- PMID: 12192032
- PMCID: PMC135624
- DOI: 10.1128/MCB.22.18.6321-6335.2002
Classification of human B-ZIP proteins based on dimerization properties
Figures





References
-
- Alber, T. 1992. Structure of the leucine zipper. Curr. Opin. Genet. Dev. 2:205-210. - PubMed
-
- Baxevanis, A. D., and C. R. Vinson. 1993. Interactions of coiled coils in transcription factors: where is the specificity? Curr. Opin. Genet. Dev. 3:278-285. - PubMed
-
- Bohmann, D., T. J. Bos, A. Admon, T. Nishimura, P. K. Vogt, and R. Tjian. 1987. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science 238:1386-1392. - PubMed
-
- Cao, Z., R. M. Umek, and S. L. McKnight. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5:1538-1552. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
