Targeted histone acetylation at the yeast CUP1 promoter requires the transcriptional activator, the TATA boxes, and the putative histone acetylase encoded by SPT10
- PMID: 12192040
- PMCID: PMC135642
- DOI: 10.1128/MCB.22.18.6406-6416.2002
Targeted histone acetylation at the yeast CUP1 promoter requires the transcriptional activator, the TATA boxes, and the putative histone acetylase encoded by SPT10
Abstract
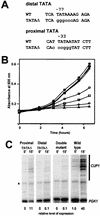
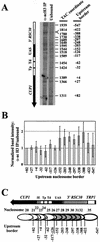
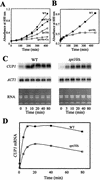
The relationship between chromatin remodeling and histone acetylation at the yeast CUP1 gene was addressed. CUP1 encodes a metallothionein required for cell growth at high copper concentrations. Induction of CUP1 with copper resulted in targeted acetylation of both H3 and H4 at the CUP1 promoter. Nucleosomes containing upstream activating sequences and sequences farther upstream were the targets for H3 acetylation. Targeted acetylation of H3 and H4 required the transcriptional activator (Ace1p) and the TATA boxes, suggesting that targeted acetylation occurs when TATA-binding protein binds to the TATA box or at a later stage in initiation. We have shown previously that induction results in nucleosome repositioning over the entire CUP1 gene, which requires Ace1p but not the TATA boxes. Therefore, the movement of nucleosomes occurring on CUP1 induction is independent of targeted acetylation. Targeted acetylation of both H3 and H4 also required the product of the SPT10 gene, which encodes a putative histone acetylase implicated in regulation at core promoters. Disruption of SPT10 was lethal at high copper concentrations and correlated with slower induction and reduced maximum levels of CUP1 mRNA. These observations constitute evidence for a novel mechanism of chromatin activation at CUP1, with a major role for the TATA box.
Figures


Similar articles
-
Remodeling of yeast CUP1 chromatin involves activator-dependent repositioning of nucleosomes over the entire gene and flanking sequences.Mol Cell Biol. 2001 Jan;21(2):534-47. doi: 10.1128/MCB.21.2.534-547.2001. Mol Cell Biol. 2001. PMID: 11134341 Free PMC article.
-
Histone H2A and Spt10 cooperate to regulate induction and autoregulation of the CUP1 metallothionein.J Biol Chem. 2005 Jan 7;280(1):104-11. doi: 10.1074/jbc.M411437200. Epub 2004 Oct 21. J Biol Chem. 2005. PMID: 15501826
-
The N-Terminal Tail of Histone H3 Regulates Copper Homeostasis in Saccharomyces cerevisiae.Mol Cell Biol. 2021 Jan 25;41(2):e00210-20. doi: 10.1128/MCB.00210-20. Print 2021 Jan 25. Mol Cell Biol. 2021. PMID: 33257505 Free PMC article.
-
TBP-associated factors in the PCAF histone acetylase complex.Cold Spring Harb Symp Quant Biol. 1998;63:493-9. doi: 10.1101/sqb.1998.63.493. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384313 Review. No abstract available.
-
Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
Cited by
-
Regulation of histone gene expression in budding yeast.Genetics. 2012 May;191(1):7-20. doi: 10.1534/genetics.112.140145. Genetics. 2012. PMID: 22555441 Free PMC article. Review.
-
Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes.Mol Cell. 2007 Mar 9;25(5):703-12. doi: 10.1016/j.molcel.2007.02.006. Epub 2007 Feb 22. Mol Cell. 2007. PMID: 17320445 Free PMC article.
-
Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae.Mol Cell Biol. 2007 Aug;27(15):5575-86. doi: 10.1128/MCB.00459-07. Epub 2007 May 25. Mol Cell Biol. 2007. PMID: 17526727 Free PMC article.
-
Global regulation by the yeast Spt10 protein is mediated through chromatin structure and the histone upstream activating sequence elements.Mol Cell Biol. 2005 Oct;25(20):9127-37. doi: 10.1128/MCB.25.20.9127-9137.2005. Mol Cell Biol. 2005. PMID: 16199888 Free PMC article.
-
A polar barrier to transcription can be circumvented by remodeler-induced nucleosome translocation.Nucleic Acids Res. 2011 May;39(9):3520-8. doi: 10.1093/nar/gkq1273. Epub 2011 Jan 17. Nucleic Acids Res. 2011. PMID: 21245049 Free PMC article.
References
-
- Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. - PubMed
-
- Berk, A. J., and P. A. Sharp. 1977. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease digest hybrids. Cell 12:721-732. - PubMed
-
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. - PubMed
-
- Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
