Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex
- PMID: 12192045
- PMCID: PMC135632
- DOI: 10.1128/MCB.22.18.6471-6479.2002
Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex
Abstract
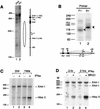
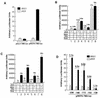
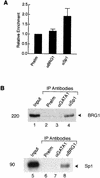
The mammalian SWI/SNF-like chromatin-remodeling BAF complex plays several important roles in controlling cell proliferation and differentiation. Interferons (IFNs) are key mediators of cellular antiviral and antiproliferative activities. In this report, we demonstrate that the BAF complex is required for the maximal induction of a subset of IFN target genes by alpha IFN (IFN-alpha). The BAF complex is constitutively associated with the IFITM3 promoter in vivo and facilitates the chromatin remodeling of the promoter upon IFN-alpha induction. Furthermore, we show that the ubiquitous transcription activator Sp1 interacts with the BAF complex in vivo and augments the BAF-mediated activation of the IFITM3 promoter. Sp1 binds constitutively to the IFITM3 promoter in the absence of the BAF complex, suggesting that it may recruit and/or stabilize the BAF complex binding to the IFITM3 promoter. Our results bring new mechanistic insights into the antiproliferative effects of the chromatin-remodeling BAF complex.
Figures

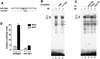

Similar articles
-
The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming.Mol Cell Biol. 2004 May;24(10):4476-86. doi: 10.1128/MCB.24.10.4476-4486.2004. Mol Cell Biol. 2004. PMID: 15121865 Free PMC article.
-
The PKR kinase promoter binds both Sp1 and Sp3, but only Sp3 functions as part of the interferon-inducible complex with ISGF-3 proteins.Virology. 2003 Sep 1;313(2):553-66. doi: 10.1016/s0042-6822(03)00347-7. Virology. 2003. PMID: 12954221
-
Activation of the RNA-dependent protein kinase PKR promoter in the absence of interferon is dependent upon Sp proteins.J Biol Chem. 2006 Feb 10;281(6):3244-53. doi: 10.1074/jbc.M510612200. Epub 2005 Dec 8. J Biol Chem. 2006. PMID: 16339759
-
Unwinding chromatin at the right places: how BAF is targeted to specific genomic locations during development.Development. 2019 Sep 30;146(19):dev178780. doi: 10.1242/dev.178780. Development. 2019. PMID: 31570369 Free PMC article. Review.
-
BAFfling pathologies: Alterations of BAF complexes in cancer.Cancer Lett. 2018 Apr 10;419:266-279. doi: 10.1016/j.canlet.2018.01.046. Epub 2018 Jan 31. Cancer Lett. 2018. PMID: 29374542 Review.
Cited by
-
Next-generation bromodomain inhibitors of the SWI/SNF complex enhance DNA damage and cell death in glioblastoma.J Cell Mol Med. 2023 Sep;27(18):2770-2781. doi: 10.1111/jcmm.17907. Epub 2023 Aug 18. J Cell Mol Med. 2023. PMID: 37593885 Free PMC article.
-
Silencing of IFN-stimulated gene transcription is regulated by histone H1 and its chaperone TAF-I.Nucleic Acids Res. 2014 Jul;42(12):7642-53. doi: 10.1093/nar/gku485. Epub 2014 May 30. Nucleic Acids Res. 2014. PMID: 24878923 Free PMC article.
-
Transcriptional control of inflammatory responses.Cold Spring Harb Perspect Biol. 2014 Sep 11;6(11):a016261. doi: 10.1101/cshperspect.a016261. Cold Spring Harb Perspect Biol. 2014. PMID: 25213094 Free PMC article. Review.
-
LncRNA XR_001779380 Primes Epithelial Cells for IFN-γ-Mediated Gene Transcription and Facilitates Age-Dependent Intestinal Antimicrobial Defense.mBio. 2021 Oct 26;12(5):e0212721. doi: 10.1128/mBio.02127-21. Epub 2021 Sep 7. mBio. 2021. PMID: 34488445 Free PMC article.
-
Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine.Cell Rep. 2021 Mar 2;34(9):108798. doi: 10.1016/j.celrep.2021.108798. Cell Rep. 2021. PMID: 33657363 Free PMC article.
References
-
- Aalfs, J. D., and R. E. Kingston. 2000. What does “chromatin remodeling” mean? Trends Biochem. Sci. 25:548-555. - PubMed
-
- Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. - PubMed
-
- Bhattacharya, S., R. Eckner, S. Grossman, E. Oldread, Z. Arany, A. D'Andrea, and D. M. Livingston. 1996. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature 383:344-347. - PubMed
-
- Boehm, U., T. Klamp, M. Groot, and J. C. Howard. 1997. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 15:749-795. - PubMed
-
- Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
