3' end processing of Drosophila melanogaster histone pre-mRNAs: requirement for phosphorylated Drosophila stem-loop binding protein and coevolution of the histone pre-mRNA processing system
- PMID: 12192062
- PMCID: PMC135633
- DOI: 10.1128/MCB.22.18.6648-6660.2002
3' end processing of Drosophila melanogaster histone pre-mRNAs: requirement for phosphorylated Drosophila stem-loop binding protein and coevolution of the histone pre-mRNA processing system
Abstract
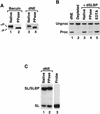
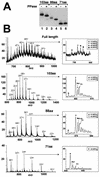
Synthetic pre-mRNAs containing the processing signals encoded by Drosophila melanogaster histone genes undergo efficient and faithful endonucleolytic cleavage in nuclear extracts prepared from Drosophila cultured cells and 0- to 13-h-old embryos. Biochemical requirements for the in vitro cleavage are similar to those previously described for the 3' end processing of mammalian histone pre-mRNAs. Drosophila 3' end processing does not require ATP and occurs in the presence of EDTA. However, in contrast to mammalian processing, Drosophila processing generates the final product ending four nucleotides after the stem-loop. Cleavage of the Drosophila substrates is abolished by depleting the extract of the Drosophila stem-loop binding protein (dSLBP), indicating that both dSLBP and the stem-loop structure in histone pre-mRNA are essential components of the processing machinery. Recombinant dSLBP expressed in insect cells by using the baculovirus system efficiently complements the depleted extract. Only the RNA-binding domain plus the 17 amino acids at the C terminus of dSLBP are required for processing. The full-length dSLBP expressed in insect cells is quantitatively phosphorylated on four residues in the C-terminal region. Dephosphorylation of the recombinant dSLBP reduces processing activity. Human and Drosophila SLBPs are not interchangeable and strongly inhibit processing in the heterologous extracts. The RNA-binding domain of the dSLBP does not substitute for the RNA-binding domain of the human SLBP in histone pre-mRNA processing in mammalian extracts. In addition to the stem-loop structure and dSLBP, 3' processing in Drosophila nuclear extracts depends on the presence of a short stretch of purines located ca. 20 nucleotides downstream from the stem, and an Sm-reactive factor, most likely the Drosophila counterpart of vertebrate U7 snRNP.
Figures



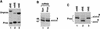
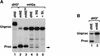

References
-
- Albig, W., and D. Doenecke. 1997. The human histone gene cluster at the D6S105 locus. Hum. Genet. 101:284-294. - PubMed
-
- Albig, W., P. Kioschis, A. Poustka, K. Meergans, and D. Doenecke. 1997. Human histone gene organization: nonregular arrangement within a large cluster. Genomics 40:314-322. - PubMed
-
- Birnstiel, M. L., and F. J. Schaufele. 1988. Structure and function of minor snRNPs, p. 155-182. In M. L. Birnstiel (ed.), Structure and function of major and minor small ribonucleoprotein particles. Springer-Verlag, Berlin, Germany.
-
- Bond, U. M., T. A. Yario, and J. A. Steitz. 1991. Multiple processing-defective mutations in a mammalian histone premessenger RNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 5:1709-1722. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
