Modern analytical ultracentrifugation in protein science: a tutorial review
- PMID: 12192063
- PMCID: PMC2373601
- DOI: 10.1110/ps.0207702
Modern analytical ultracentrifugation in protein science: a tutorial review
Abstract
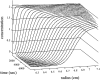
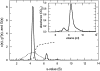
Analytical ultracentrifugation (AU) is reemerging as a versatile tool for the study of proteins. Monitoring the sedimentation of macromolecules in the centrifugal field allows their hydrodynamic and thermodynamic characterization in solution, without interaction with any matrix or surface. The combination of new instrumentation and powerful computational software for data analysis has led to major advances in the characterization of proteins and protein complexes. The pace of new advancements makes it difficult for protein scientists to gain sufficient expertise to apply modern AU to their research problems. To address this problem, this review builds from the basic concepts to advanced approaches for the characterization of protein systems, and key computational and internet resources are provided. We will first explore the characterization of proteins by sedimentation velocity (SV). Determination of sedimentation coefficients allows for the modeling of the hydrodynamic shape of proteins and protein complexes. The computational treatment of SV data to resolve sedimenting components has been achieved. Hence, SV can be very useful in the identification of the oligomeric state and the stoichiometry of heterogeneous interactions. The second major part of the review covers sedimentation equilibrium (SE) of proteins, including membrane proteins and glycoproteins. This is the method of choice for molar mass determinations and the study of self-association and heterogeneous interactions, such as protein-protein, protein-nucleic acid, and protein-small molecule binding.
Figures
Comment in
-
Modern analytical ultracentrifugation in protein science: look forward, not back.Protein Sci. 2003 Nov;12(11):2647-9; discussion 2649-50. doi: 10.1110/ps.0235803. Protein Sci. 2003. PMID: 14573877 Free PMC article.
References
-
- Ansevin, A.T., Roark, D.E., and Yphantis, D.A. 1970. Improved ultracentrifuge cells for high-speed sedimentation equilibrium studies with interference optics. Anal. Biochem. 34 237–261. - PubMed
-
- Arkin, M. and Lear, J.D. 2001. A new data analysis method to determine binding constants of small molecules using equilibrium analytical ultracentrifugation with absorption optics. Anal. Biochem. 299 98–107. - PubMed
-
- Arthos, J., Cicala, C., Steenbeke, T.D., VanRyk, D., Dela Cruz, C., Khazanie, P., Selig, S.M., Hanback, D.B., Nam, D., Schuck, P., et al. 2002. Efficient inhibition of HIV-1 viral replication by a novel modification of sCD4. J. Biol. Chem. 277 11456–11464. - PubMed
-
- Bailey, M.F., Davidson, B.E., Minton, A.P., Sawyer, W.H., and Howlett, G.J. 1996. The effect of self-association on the interaction of the Escherichia coli regulatory protein TyrR with DNA. J. Mol. Biol. 263 671–684. - PubMed
-
- Becerra, S.P., Kumar, A., Lewis, M.S., Widen, S.G., Abbots, J., Karawya, E.M., Hughes, S.H., Shiloach, J., and Wilson, S.H. 1991. Protein–protein interactions of HIV-1 reverse transcriptase: Implication of central and C-terminal regions in subunit binding. (Lewis, MS. Appendix: Ultracentrifuge analysis of a mixed association.). Biochemistry 30 11707–11719. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

