The effect of prime-site occupancy on the hepatitis C virus NS3 protease structure
- PMID: 12192066
- PMCID: PMC2373603
- DOI: 10.1110/ps.0206602
The effect of prime-site occupancy on the hepatitis C virus NS3 protease structure
Abstract
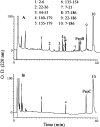
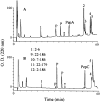
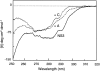
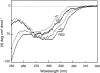
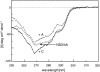
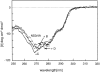
We recently reported a new class of inhibitors of the chymotrypsin-like serine protease NS3 of the hepatitis C virus. These inhibitors exploit the binding potential of the S' site of the protease, which is not generally used by the natural substrates. The effect of prime-site occupancy was analyzed by circular dichroism spectroscopy and limited proteolysis-mass spectrometry. Generally, nonprime inhibitors cause a structural change in NS3. Binding in the S' site produces additional conformational changes with different binding modes, even in the case of the NS3/4A cofactor complex. Notably, inhibitor binding either in the S or S' site also has profound effects on the stabilization of the protease. In addition, the stabilization propagates to regions not in direct contact with the inhibitor. In particular, the N-terminal region, which according to structural studies is endowed with low structural stability and is not stabilized by nonprime inhibitors, was now fully protected from proteolytic degradation. From the perspective of drug design, P-P' inhibitors take advantage of binding pockets, which are not exploited by the natural HCV substrates; hence, they are an entry point for a novel class of NS3/4A inhibitors. Here we show that binding of each inhibitor is associated with a specific structural rearrangement. The development of a range of inhibitors belonging to different classes and an understanding of their interactions with the protease are required to address the issue of the most likely outcome of viral protease inhibitor therapy, that is, viral resistance.
Figures
Similar articles
-
Conformational changes in human hepatitis C virus NS3 protease upon binding of product-based inhibitors.Biochemistry. 1999 Oct 19;38(42):13844-52. doi: 10.1021/bi991220w. Biochemistry. 1999. PMID: 10529230
-
Mechanistic role of an NS4A peptide cofactor with the truncated NS3 protease of hepatitis C virus: elucidation of the NS4A stimulatory effect via kinetic analysis and inhibitor mapping.Biochemistry. 1997 Aug 5;36(31):9340-8. doi: 10.1021/bi963054n. Biochemistry. 1997. PMID: 9235976
-
Prime site binding inhibitors of a serine protease: NS3/4A of hepatitis C virus.Biochemistry. 2002 Apr 30;41(17):5483-92. doi: 10.1021/bi025603x. Biochemistry. 2002. PMID: 11969409
-
Hepatitis C virus NS3/4A protease.Antiviral Res. 1999 Feb;41(1):67-84. Antiviral Res. 1999. PMID: 10321580 Review.
-
The hepatitis C virus NS3 proteinase: structure and function of a zinc-containing serine proteinase.Antivir Ther. 1998;3(Suppl 3):99-109. Antivir Ther. 1998. PMID: 10726060 Review.
Cited by
-
New therapies for chronic hepatitis C virus infection.Curr Gastroenterol Rep. 2004 Feb;6(1):77-86. doi: 10.1007/s11894-004-0030-5. Curr Gastroenterol Rep. 2004. PMID: 14720458 Review.
-
Mass Spectrometry-Based Proteomics Technologies to Define Endogenous Protein-Protein Interactions and Their Applications to Cancer and Viral Infectious Diseases.Mass Spectrom Rev. 2025 Feb 9:10.1002/mas.21926. doi: 10.1002/mas.21926. Online ahead of print. Mass Spectrom Rev. 2025. PMID: 39924651 Free PMC article. Review.
-
A lead discovery strategy driven by a comprehensive analysis of proteases in the peptide substrate space.Protein Sci. 2010 Nov;19(11):2096-109. doi: 10.1002/pro.490. Protein Sci. 2010. PMID: 20799349 Free PMC article.
References
-
- Atherton, E. and Sheppard, R.C. 1989. Solid-phase peptide synthesis, a practical approach, IRL Press, Oxford.
-
- Atkinson, R.A., Joseph, C., Dal Piaz, F., Birolo, L., Stier, G., Pucci, P., and Pastore, A. 2000. Binding of α-actinin to titin: Implications for Z-disk assembly. Biochemistry 39 5255–5264. - PubMed
-
- Barbato, G., Cicero, D.O., Nardi, M.C., Steinkühler, C., Cortese, R., De Francesco, R., and Bazzo, R. 1999. The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism. J. Mol. Biol. 289 371–384. - PubMed
-
- Bianchi, E., Urbani, A., Biasiol, G., Brunetti, M., Pessi, A., De Francesco, R., and Steinkühler, C. 1997. Complex formation between the hepatitis C virus serine proteinase and a synthetic NS4A cofactor peptide. Biochemistry 36 7890–7897. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

