A structurally conserved water molecule in Rossmann dinucleotide-binding domains
- PMID: 12192068
- PMCID: PMC2373605
- DOI: 10.1110/ps.0213502
A structurally conserved water molecule in Rossmann dinucleotide-binding domains
Abstract
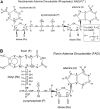
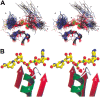
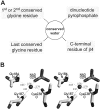
A computational comparison of 102 high-resolution (</=1.90 A) enzyme-dinucleotide (NAD, NADP, FAD) complexes was performed to investigate the role of solvent in dinucleotide recognition by Rossmann fold domains. The typical binding site contains about 9-12 water molecules, and about 30% of the hydrogen bonds between the protein and the dinucleotide are water mediated. Detailed inspection of the structures reveals a structurally conserved water molecule bridging dinucleotides with the well-known glycine-rich phosphate-binding loop. This water molecule displays a conserved hydrogen-bonding pattern. It forms hydrogen bonds to the dinucleotide pyrophosphate, two of the three conserved glycine residues of the phosphate-binding loop, and a residue at the C-terminus of strand four of the Rossmann fold. The conserved water molecule is also present in high-resolution structures of apo enzymes. However, the water molecule is not present in structures displaying significant deviations from the classic Rossmann fold motif, such as having nonstandard topology, containing a very short phosphate-binding loop, or having alpha-helix "A" oriented perpendicular to the beta-sheet. Thus, the conserved water molecule appears to be an inherent structural feature of the classic Rossmann dinucleotide-binding domain.
Figures

Similar articles
-
GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding Rossmann folds through C(alpha)-H... O hydrogen bonds and van der waals interactions.J Mol Biol. 2002 Oct 11;323(1):69-76. doi: 10.1016/s0022-2836(02)00885-9. J Mol Biol. 2002. PMID: 12368099
-
Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate.J Mol Biol. 2008 Jul 11;380(3):532-47. doi: 10.1016/j.jmb.2008.04.072. Epub 2008 May 7. J Mol Biol. 2008. PMID: 18565343
-
FAD-binding site and NADP reactivity in human renalase: a new enzyme involved in blood pressure regulation.J Mol Biol. 2011 Aug 12;411(2):463-73. doi: 10.1016/j.jmb.2011.06.010. Epub 2011 Jun 14. J Mol Biol. 2011. PMID: 21699903
-
NAD-binding domains of dehydrogenases.Curr Opin Struct Biol. 1995 Dec;5(6):775-83. doi: 10.1016/0959-440x(95)80010-7. Curr Opin Struct Biol. 1995. PMID: 8749365 Review.
-
The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins.FASEB J. 1996 Sep;10(11):1257-69. doi: 10.1096/fasebj.10.11.8836039. FASEB J. 1996. PMID: 8836039 Review.
Cited by
-
Characterization of a HoxEFUYH type of [NiFe] hydrogenase from Allochromatium vinosum and some EPR and IR properties of the hydrogenase module.J Biol Inorg Chem. 2007 Jan;12(1):62-78. doi: 10.1007/s00775-006-0162-1. Epub 2006 Sep 13. J Biol Inorg Chem. 2007. PMID: 16969669
-
Proanthocyanidin Synthesis in Chinese Bayberry (Myrica rubra Sieb. et Zucc.) Fruits.Front Plant Sci. 2018 Feb 28;9:212. doi: 10.3389/fpls.2018.00212. eCollection 2018. Front Plant Sci. 2018. PMID: 29541082 Free PMC article.
-
Yeast alcohol dehydrogenase structure and catalysis.Biochemistry. 2014 Sep 16;53(36):5791-803. doi: 10.1021/bi5006442. Epub 2014 Sep 3. Biochemistry. 2014. PMID: 25157460 Free PMC article.
-
Targeting Penicillium expansum GMC Oxidoreductase with High Affinity Small Molecules for Reducing Patulin Production.Biology (Basel). 2020 Dec 31;10(1):21. doi: 10.3390/biology10010021. Biology (Basel). 2020. PMID: 33396459 Free PMC article.
-
Water potential governs the effector specificity of the transcriptional regulator XylR of Pseudomonas putida.Environ Microbiol. 2023 May;25(5):1041-1054. doi: 10.1111/1462-2920.16342. Epub 2023 Jan 31. Environ Microbiol. 2023. PMID: 36683138 Free PMC article.
References
-
- Adolph, H.W., Zwart, P., Meijers, R., Hubatsch, I., Kiefer, M., Lamzin, V., and Cedergren-Zeppezauer, E. 2000. Structural basis for substrate specificity differences of horse liver alcohol dehydrogenase isozymes. Biochemistry 39 12885–12897. - PubMed
-
- Al-Karadaghi, S., Cedergren-Zeppezauer, E.S., Petratos, K., Hovmoeller, S., Terry, H., Dauter, Z., and Wilson, K.S. 1994. Refined crystal structure of liver alcohol dehydrogenase-NADH complex at 1.8 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 50 793–807. - PubMed
-
- Allaire, M., Li, Y., MacKenzie, R.E., and Cygler, M. 1998. The 3-D structure of a folate-dependent dehydrogenase/cyclohydrolase bifunctional enzyme at 1.5 Å resolution. Structure 6 173–182. - PubMed
-
- Aronov, A.M., Verlinde, C.L., Hol, W.G., and Gelb, M.H. 1998. Selective tight binding inhibitors of trypanosomal glyceraldehyde-3-phosphate dehydrogenase via structure-based drug design. J. Med. Chem. 41 4790–4799. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

