The beta-barrel finder (BBF) program, allowing identification of outer membrane beta-barrel proteins encoded within prokaryotic genomes
- PMID: 12192075
- PMCID: PMC2373602
- DOI: 10.1110/ps.0209002
The beta-barrel finder (BBF) program, allowing identification of outer membrane beta-barrel proteins encoded within prokaryotic genomes
Abstract
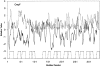
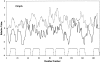
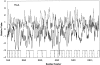
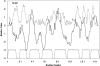
Many outer membrane proteins (OMPs) in Gram-negative bacteria possess known beta-barrel three-dimensional (3D) structures. These proteins, including channel-forming transmembrane porins, are diverse in sequence but exhibit common structural features. We here report computational analyses of six outer membrane proteins of known 3D structures with respect to (1) secondary structure, (2) hydropathy, and (3) amphipathicity. Using these characteristics, as well as the presence of an N-terminal targeting sequence, a program was developed allowing prediction of integral membrane beta-barrel proteins encoded within any completely sequenced prokaryotic genome. This program, termed the beta-barrel finder (BBF) program, was used to analyze the proteins encoded within the Escherichia coli genome. Out of 4290 sequences examined, 118 (2.8%) were retrieved. Of these, almost all known outer membrane proteins with established beta-barrel structures as well as many probable outer membrane proteins were identified. This program should be useful for predicting the occurrence of outer membrane proteins in bacteria with completely sequenced genomes.
Figures

References
-
- Achouak, W., Heulin, T., and Pagés, J.-M. 2001. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 199 1–7. - PubMed
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Arora, A., Rinehart, D., Szabo, G., and Tamm, L.K. 2000. Refolded outer membrane protein A of Escherichia coli forms ion channels with two conductance states in planar lipid bilayers. J. Biol. Chem. 275 1594–1600. - PubMed
-
- Arrecubieta, C., Hammarton, T.C., Barrett, B., Chareonsudjai, S., Hodson, N., Rainey, D., and Roberts, I.S. 2001. The transport of group 2 capsular polysaccharides across the periplasmic space in Escherichia coli. J. Biol. Chem. 276 4245–4250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

