Mapping sequence differences between thimet oligopeptidase and neurolysin implicates key residues in substrate recognition
- PMID: 12192079
- PMCID: PMC2373592
- DOI: 10.1110/ps.0216302
Mapping sequence differences between thimet oligopeptidase and neurolysin implicates key residues in substrate recognition
Abstract
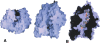
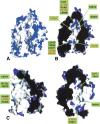
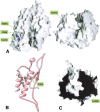
The highly homologous endopeptidases thimet oligopeptidase and neurolysin are both restricted to short peptide substrates and share many of the same cleavage sites on bioactive and synthetic peptides. They sometimes target different sites on the same peptide, however, and defining the determinants of differential recognition will help us to understand how both enzymes specifically target a wide variety of cleavage site sequences. We have mapped the positions of the 224 surface residues that differ in sequence between the two enzymes onto the surface of the neurolysin crystal structure. Although the deep active site channel accounts for about one quarter of the total surface area, only 11% of the residue differences map to this region. Four isolated sequence changes (R470/E469, R491/M490, N496/H495, and T499/R498; neurolysin residues given first) are well positioned to affect recognition of substrate peptides, and differences in cleavage site specificity can be largely rationalized on the basis of these changes. We also mapped the positions of three cysteine residues believed to be responsible for multimerization of thimet oligopeptidase, a process that inactivates the enzyme. These residues are clustered on the outside of one channel wall, where multimerization via disulfide formation is unlikely to block the substrate-binding site. Finally, we mapped the regulatory phosphorylation site in thimet oligopeptidase to a location on the outside of the molecule well away from the active site, which indicates this modification has an indirect effect on activity.
Figures



Similar articles
-
Swapping the substrate specificities of the neuropeptidases neurolysin and thimet oligopeptidase.J Biol Chem. 2007 Mar 30;282(13):9722-9732. doi: 10.1074/jbc.M609897200. Epub 2007 Jan 24. J Biol Chem. 2007. PMID: 17251185
-
Crystal structure of human thimet oligopeptidase provides insight into substrate recognition, regulation, and localization.J Biol Chem. 2004 May 7;279(19):20480-9. doi: 10.1074/jbc.M400795200. Epub 2004 Mar 3. J Biol Chem. 2004. PMID: 14998993
-
Characterization of a mitochondrial metallopeptidase reveals neurolysin as a homologue of thimet oligopeptidase.J Biol Chem. 1995 Feb 3;270(5):2092-8. doi: 10.1074/jbc.270.5.2092. J Biol Chem. 1995. PMID: 7836437
-
Soluble metalloendopeptidases and neuroendocrine signaling.Endocr Rev. 2002 Oct;23(5):647-64. doi: 10.1210/er.2001-0032. Endocr Rev. 2002. PMID: 12372844 Review.
-
Thimet oligopeptidase--a review of a thiol dependent metallo-endopeptidase also known as Pz-peptidase endopeptidase 24.15 and endo-oligopeptidase.Biol Chem Hoppe Seyler. 1993 Feb;374(2):91-100. Biol Chem Hoppe Seyler. 1993. PMID: 8471182 Review.
Cited by
-
Neurolysin Knockout Mice in a Diet-Induced Obesity Model.Int J Mol Sci. 2023 Oct 14;24(20):15190. doi: 10.3390/ijms242015190. Int J Mol Sci. 2023. PMID: 37894869 Free PMC article.
-
The Relevance of Thimet Oligopeptidase in the Regulation of Energy Metabolism and Diet-Induced Obesity.Biomolecules. 2020 Feb 17;10(2):321. doi: 10.3390/biom10020321. Biomolecules. 2020. PMID: 32079362 Free PMC article.
-
Development of fluorogenic substrates for colorectal tumor-related neuropeptidases for activity-based diagnosis.Chem Sci. 2023 Apr 11;14(17):4495-4499. doi: 10.1039/d2sc07029d. eCollection 2023 May 3. Chem Sci. 2023. PMID: 37152255 Free PMC article.
-
EP24.15 as a Potential Regulator of Kisspeptin Within the Neuroendocrine Hypothalamus.Endocrinology. 2016 Feb;157(2):820-30. doi: 10.1210/en.2015-1580. Epub 2015 Dec 11. Endocrinology. 2016. PMID: 26653570 Free PMC article.
-
Characterization of thimet oligopeptidase and neurolysin activities in B16F10-Nex2 tumor cells and their involvement in angiogenesis and tumor growth.Mol Cancer. 2007 Jul 9;6:44. doi: 10.1186/1476-4598-6-44. Mol Cancer. 2007. PMID: 17620116 Free PMC article.
References
-
- Achstetter, T., Ehmann, C., Osaki, A., and Wolf, D.H. 1984. Proteolysis in eukaryotic cells: Proteinase yscE, a new yeast peptidase. J. Biol. Chem. 259 13344–13348. - PubMed
-
- Achstetter, T., Ehmann, C., and Wolf, D.H. 1985. Proteinase yscD: Purification and characterization of a new yeast peptidase. J. Biol. Chem. 260 4585–4590. - PubMed
-
- Adam, G., and Delbrück, M. 1968. Reduction of dimensionality in biological diffusion processes. In Structural chemistry and molecular biology. (eds. A. Rich and N. Davidson) pp. 198–207. Freeman, San Francisco.
-
- Barrett, A.J., Brown, M.A., Dando, P.M., Knight, C.G., McKie, N., Rawlings, N.D., and Serizawa, A. 1995. Thimet oligopeptidase and oligopeptidase M or neurolysin. Meth. Enzymol. 248 529–556. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

