A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25-27
- PMID: 12192642
- PMCID: PMC378535
- DOI: 10.1086/342720
A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25-27
Abstract
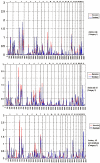
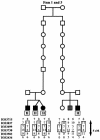
To identify genetic loci for autism-spectrum disorders, we have performed a two-stage genomewide scan in 38 Finnish families. The detailed clinical examination of all family members revealed infantile autism, but also Asperger syndrome (AS) and developmental dysphasia, in the same set of families. The most significant evidence for linkage was found on chromosome 3q25-27, with a maximum two-point LOD score of 4.31 (Z(max )(dom)) for D3S3037, using infantile autism and AS as an affection status. Six markers flanking over a 5-cM region on 3q gave Z(max dom) >3, and a maximum parametric multipoint LOD score (MLS) of 4.81 was obtained in the vicinity of D3S3715 and D3S3037. Association, linkage disequilibrium, and haplotype analyses provided some evidence for shared ancestor alleles on this chromosomal region among affected individuals, especially in the regional subisolate. Additional potential susceptibility loci with two-point LOD scores >2 were observed on chromosomes 1q21-22 and 7q. The region on 1q21-22 overlaps with the previously reported candidate region for infantile autism and schizophrenia, whereas the region on chromosome 7q provided evidence for linkage 58 cM distally from the previously described autism susceptibility locus (AUTS1).
Figures



References
Electronic-Database Information
-
- Généthon, http://www.genethon.fr/php/index.php
-
- Genetic Location Database, http://cedar.genetics.soton.ac.uk/public_html/ldb.html
-
- Marshfield, http://research.marshfieldclinic.org/genetics/
-
- NCBI, http://www.ncbi.nlm.nih.gov/ (for draft sequence used to order markers)
References
-
- Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA (1999) Genetic studies of autistic disorder and chromosome 7. Genomics 61:227–236 - PubMed
-
- Auranen M, Nieminen T, Majuri S, Vanhala R, Peltonen L, Järvelä I (2000) Analysis of autism susceptiblity gene loci on chromosomes 1p, 4p, 6q, 7q, 13q, 15q, 16p, 17q, 19q and 22q in Finnish multiplex families. Mol Psychiatry 5:320–322 - PubMed
-
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25:63–77 - PubMed
-
- Bailey A, Palferman S, Heavey L, Le Couteur A (1998) Autism: the phenotype in relatives. J Autism Dev Disord 28:369–392 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

