Mycobacterium smegmatis L-alanine dehydrogenase (Ald) is required for proficient utilization of alanine as a sole nitrogen source and sustained anaerobic growth
- PMID: 12193615
- PMCID: PMC135311
- DOI: 10.1128/JB.184.18.5001-5010.2002
Mycobacterium smegmatis L-alanine dehydrogenase (Ald) is required for proficient utilization of alanine as a sole nitrogen source and sustained anaerobic growth
Abstract
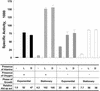
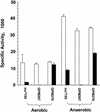
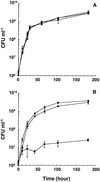
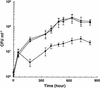
NAD(H)-dependent L-alanine dehydrogenase (EC 1.4.1.1) (Ald) catalyzes the oxidative deamination of L-alanine and the reductive amination of pyruvate. To assess the physiological role of Ald in Mycobacterium smegmatis, we cloned the ald gene, identified its promoter, determined the protein expression levels, and analyzed the combined effects of nutrient supplementation, oxygen availability, and growth stage on enzyme activity. High Ald activities were observed in cells grown in the presence of L- or D-alanine regardless of the oxygen availability and growth stage. In exponentially growing cells under aerobic conditions, supplementation with alanine resulted in a 25- to 50-fold increase in the enzyme activity. In the absence of alanine supplementation, 23-fold-higher Ald activities were observed in cells grown exponentially under anaerobic conditions. Furthermore, M. smegmatis ald null mutants were constructed by targeted disruption and were shown to lack any detectable Ald activity. In contrast, the glycine dehydrogenase (EC 1.4.1.10) (Gdh) activity in mutant cells remained at wild-type levels, indicating that another enzyme protein is responsible for the physiologically relevant reductive amination of glyoxylate. The ald mutants grew poorly in minimal medium with L-alanine as the sole nitrogen source, reaching a saturation density 100-fold less than that of the wild-type strain. Likewise, mutants grew to a saturation density 10-fold less than that of the wild-type strain under anaerobic conditions. In summary, the phenotypes displayed by the M. smegmatis ald mutants suggest that Ald plays an important role in both alanine utilization and anaerobic growth.
Figures

References
-
- Allaway, D., E. M. Lodwig, L. A. Crompton, M. Wood, R. Parsons, T. R. Wheeler, and P. S. Poole. 2000. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36:508-515. - PubMed
-
- Bashyam, M. D., and A. Tyagi. 1994. An efficient and high-yielding method for isolation of RNA from mycobacteria. BioTechniques 17:834-836. - PubMed
-
- Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

