Cloning the vaccinia virus genome as a bacterial artificial chromosome in Escherichia coli and recovery of infectious virus in mammalian cells
- PMID: 12196634
- PMCID: PMC129459
- DOI: 10.1073/pnas.192420599
Cloning the vaccinia virus genome as a bacterial artificial chromosome in Escherichia coli and recovery of infectious virus in mammalian cells
Abstract
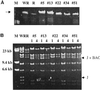
The ability to manipulate the vaccinia virus (VAC) genome, as a plasmid in bacteria, would greatly facilitate genetic studies and provide a powerful alternative method of making recombinant viruses. VAC, like other poxviruses, has a linear, double-stranded DNA genome with covalently closed hairpin ends that are resolved from transient head-to-head and tail-to-tail concatemers during replication in the cytoplasm of infected cells. Our strategy to construct a nearly 200,000-bp VAC-bacterial artificial chromosome (BAC) was based on circularization of head-to-tail concatemers of VAC DNA. Cells were infected with a recombinant VAC containing inserted sequences for plasmid replication and maintenance in Escherichia coli; DNA concatemer resolution was inhibited leading to formation and accumulation of head-to-tail concatemers, in addition to the usual head-to-head and tail-to-tail forms; the concatemers were circularized by homologous or Cre-loxP-mediated recombination; and E. coli were transformed with DNA from the infected cell lysates. Stable plasmids containing the entire VAC genome, with an intact concatemer junction sequence, were identified. Rescue of infectious VAC was consistently achieved by transfecting the VAC-BAC plasmids into mammalian cells that were infected with a helper nonreplicating fowlpox virus.
Figures




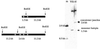
Similar articles
-
Genetic manipulation of poxviruses using bacterial artificial chromosome recombineering.Methods Mol Biol. 2012;890:37-57. doi: 10.1007/978-1-61779-876-4_3. Methods Mol Biol. 2012. PMID: 22688760
-
Engineering of a vaccinia virus bacterial artificial chromosome in Escherichia coli by bacteriophage lambda-based recombination.Nat Methods. 2005 Feb;2(2):95-7. doi: 10.1038/nmeth734. Nat Methods. 2005. PMID: 15782205
-
Molecular cloning of the guinea pig cytomegalovirus (GPCMV) genome as an infectious bacterial artificial chromosome (BAC) in Escherichia coli.Mol Genet Metab. 2001 Jan;72(1):15-26. doi: 10.1006/mgme.2000.3102. Mol Genet Metab. 2001. PMID: 11161824
-
Cloning of herpesviral genomes as bacterial artificial chromosomes.Rev Med Virol. 2003 Mar-Apr;13(2):111-21. doi: 10.1002/rmv.380. Rev Med Virol. 2003. PMID: 12627394 Review.
-
Recent advances in herpesvirus genetics using bacterial artificial chromosomes.Mol Genet Metab. 2001 Jan;72(1):8-14. doi: 10.1006/mgme.2000.3123. Mol Genet Metab. 2001. PMID: 11161823 Review.
Cited by
-
Expression and cellular immunogenicity of a transgenic antigen driven by endogenous poxviral early promoters at their authentic loci in MVA.PLoS One. 2012;7(6):e40167. doi: 10.1371/journal.pone.0040167. Epub 2012 Jun 27. PLoS One. 2012. PMID: 22761956 Free PMC article.
-
Concurrent expression of HP-NAP enhances antitumor efficacy of oncolytic vaccinia virus but not for Semliki Forest virus.Mol Ther Oncolytics. 2021 May 5;21:356-366. doi: 10.1016/j.omto.2021.04.016. eCollection 2021 Jun 25. Mol Ther Oncolytics. 2021. PMID: 34141872 Free PMC article.
-
Synthetic horsepox viruses and the continuing debate about dual use research.PLoS Pathog. 2018 Oct 4;14(10):e1007025. doi: 10.1371/journal.ppat.1007025. eCollection 2018 Oct. PLoS Pathog. 2018. PMID: 30286190 Free PMC article. No abstract available.
-
Vaccinia virus G9 protein is an essential component of the poxvirus entry-fusion complex.J Virol. 2006 Oct;80(19):9822-30. doi: 10.1128/JVI.00987-06. J Virol. 2006. PMID: 16973586 Free PMC article.
-
Killing a killer: what next for smallpox?PLoS Pathog. 2010 Jan 29;6(1):e1000727. doi: 10.1371/journal.ppat.1000727. PLoS Pathog. 2010. PMID: 20126444 Free PMC article. Review. No abstract available.
References
-
- Moss B. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), Vol. 2, pp. 2849–2883.
-
- Fenner F., Henderson, D. A., Arita, I., Jezek, Z. & Ladnyi, I. D., (1988) Smallpox and its Eradication (World Health Organization, Geneva).
-
- Earl P. L., Moss, B., Wyatt, L. S. & Carroll, M. W. (1998) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Greene & Wiley, New York), Vol. 2, pp. 16.17.1–16.17.19.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

