Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA-targeted probes
- PMID: 12200269
- PMCID: PMC124080
- DOI: 10.1128/AEM.68.9.4225-4232.2002
Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA-targeted probes
Abstract
A 16S rRNA-targeted probe was designed and validated in order to quantify the number of uncultured Ruminococcus obeum-like bacteria by fluorescent in situ hybridization (FISH). These bacteria have frequently been found in 16S ribosomal DNA clone libraries prepared from bacterial communities in the human intestine. Thirty-two reference strains from the human intestine, including a phylogenetically related strain and strains of some other Ruminococcus species, were used as negative controls and did not hybridize with the new probe. Microscopic and flow cytometric analyses revealed that a group of morphologically similar bacteria in feces did hybridize with this probe. Moreover, it was found that all hybridizing cells also hybridized with a probe specific for the Clostridium coccoides-Eubacterium rectale group, a group that includes the uncultured R. obeum-like bacteria. Quantification of the uncultured R. obeum-like bacteria and the C. coccoides-E. rectale group by flow cytometry and microscopy revealed that these groups comprised approximately 2.5 and 16% of the total community in fecal samples, respectively. The uncultured R. obeum-like bacteria comprise about 16% of the C. coccoides-E. rectale group. These results indicate that the uncultured R. obeum-like bacteria are numerically important in human feces. Statistical analysis revealed no significant difference between the microscopic and flow cytometric counts and the different feces sampling times, while a significant host-specific effect on the counts was observed. Our data demonstrate that the combination of FISH and flow cytometry is a useful approach for studying the ecology of uncultured bacteria in the human gastrointestinal tract.
Figures


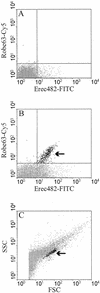
References
-
- Bry, L., P. G. Falk, T. Midtvedt, and J. I. Gordon. 1996. A model of host-microbial interactions in an open mammalian ecosystem. Science 273:1381-1383. - PubMed
-
- Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

