Quantitative tracing, by Taq nuclease assays, of a synechococcus ecotype in a highly diversified natural population
- PMID: 12200304
- PMCID: PMC124134
- DOI: 10.1128/AEM.68.9.4486-4494.2002
Quantitative tracing, by Taq nuclease assays, of a synechococcus ecotype in a highly diversified natural population
Abstract
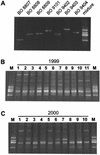
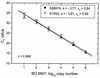
Quantitative Taq nuclease assays (TNAs) (TaqMan PCR), nested PCR in combination with denaturing gradient gel electrophoresis (DGGE), and epifluorescence microscopy were used to analyze the autotrophic picoplankton (APP) of Lake Constance. Microscopic analysis revealed dominance of phycoerythrin (PE)-rich Synechococcus spp. in the pelagic zone of this lake. Cells passing a 3- micro m-pore-size filter were collected during the growth period of the years 1999 and 2000. The diversity of PE-rich Synechococcus spp. was examined using DGGE to analyze GC-clamped amplicons of a noncoding section of the 16S-23S intergenic spacer in the ribosomal operon. In both years, genotypes represented by three closely related PE-rich Synechococcus strains of our culture collection dominated the population, while other isolates were traced sporadically or were not detected in their original habitat by this method. For TNAs, primer-probe combinations for two taxonomic levels were used, one to quantify genomes of all known Synechococcus-type cyanobacteria in the APP of Lake Constance and one to enumerate genomes of a single ecotype represented by the PE-rich isolate Synechococcus sp. strain BO 8807. During the growth period, genome numbers of known Synechococcus spp. varied by 2 orders of magnitude (2.9 x 10(3) to 3.1 x 10(5) genomes per ml). The ecotype Synechococcus sp. strain BO 8807 was detected in every sample at concentrations between 1.6 x 10(1) and 1.3 x 10(4) genomes per ml, contributing 0.02 to 5.7% of the quantified cyanobacterial picoplankton. Although the quantitative approach taken in this study has disclosed several shortcomings in the sampling and detection methods, this study demonstrated for the first time the extensive internal dynamics that lie beneath the seemingly arbitrary variations of a population of microbial photoautotrophs in the pelagic habitat.
Figures

References
-
- Assmann, D. 1998. Nahrungsselektion und Nahrungsverwertung Chroococcaler Cyanobakterien durch Heterotrophe Nanoflagellaten. Ph.D. thesis. Universität Konstanz, Konstanz, Germany.
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
-
- Ernst, A. 1991. Cyanobacterial picoplankton from Lake Constance. I. Isolation by fluorescence characteristics. J. Plankton Res. 13:1307-1312.
-
- Ernst, A., S. Becker, K. Hennes, and C. Postius. 2000. Is there a succession in the autotrophic picoplankton of temperate zone lakes?, p. 623-629. In C. R. Bell, M. Brylinski, and P. Johnson-Green (ed.), Microbial biosystems: new frontiers. Proceedings of the 8th International Symposium on Microbial Ecology. Atlantic Canada Society for Microbial Ecology, Halifax, Canada.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

