Enzymatic degradation of chlorodiamino-s-triazine
- PMID: 12200330
- PMCID: PMC124118
- DOI: 10.1128/AEM.68.9.4672-4675.2002
Enzymatic degradation of chlorodiamino-s-triazine
Abstract
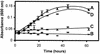
2-Chloro-4,6-diamino-s-triazine (CAAT) is a metabolite of atrazine biodegradation in soils. Atrazine chlorohydrolase (AtzA) catalyzes the dechlorination of atrazine but is unreactive with CAAT. In this study, melamine deaminase (TriA), which is 98% identical to AtzA, catalyzed deamination of CAAT to produce 2-chloro-4-amino-6-hydroxy-s-triazine (CAOT). CAOT underwent dechlorination via hydroxyatrazine ethylaminohydrolase (AtzB) to yield ammelide. This represents a newly discovered dechlorination reaction for AtzB. Ammelide was subsequently hydrolyzed by N-isopropylammelide isopropylaminohydrolase to produce cyanuric acid, a compound metabolized by a variety of soil bacteria.
Figures
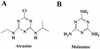
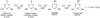
Similar articles
-
Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different.J Bacteriol. 2001 Apr;183(8):2405-10. doi: 10.1128/JB.183.8.2405-2410.2001. J Bacteriol. 2001. PMID: 11274097 Free PMC article.
-
Hydroxyatrazine N-ethylaminohydrolase (AtzB): an amidohydrolase superfamily enzyme catalyzing deamination and dechlorination.J Bacteriol. 2007 Oct;189(19):6989-97. doi: 10.1128/JB.00630-07. Epub 2007 Jul 27. J Bacteriol. 2007. PMID: 17660279 Free PMC article.
-
Biodegradation of atrazine and related s-triazine compounds: from enzymes to field studies.Appl Microbiol Biotechnol. 2002 Jan;58(1):39-45. doi: 10.1007/s00253-001-0862-y. Appl Microbiol Biotechnol. 2002. PMID: 11831474 Review.
-
AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes.J Bacteriol. 1998 Jan;180(1):152-8. doi: 10.1128/JB.180.1.152-158.1998. J Bacteriol. 1998. PMID: 9422605 Free PMC article.
-
Bacterial catabolism of s-triazine herbicides: biochemistry, evolution and application.Adv Microb Physiol. 2020;76:129-186. doi: 10.1016/bs.ampbs.2020.01.004. Epub 2020 Feb 11. Adv Microb Physiol. 2020. PMID: 32408946 Review.
Cited by
-
Functional Trade-Offs in Promiscuous Enzymes Cannot Be Explained by Intrinsic Mutational Robustness of the Native Activity.PLoS Genet. 2016 Oct 7;12(10):e1006305. doi: 10.1371/journal.pgen.1006305. eCollection 2016 Oct. PLoS Genet. 2016. PMID: 27716796 Free PMC article.
-
Evolution of catabolic pathways: Genomic insights into microbial s-triazine metabolism.J Bacteriol. 2007 Feb;189(3):674-82. doi: 10.1128/JB.01257-06. Epub 2006 Nov 17. J Bacteriol. 2007. PMID: 17114259 Free PMC article. Review. No abstract available.
-
Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.PLoS One. 2019 Jun 10;14(6):e0216979. doi: 10.1371/journal.pone.0216979. eCollection 2019. PLoS One. 2019. PMID: 31181074 Free PMC article.
-
Phylogenetics in the bioinformatics culture of understanding.Comp Funct Genomics. 2004;5(2):128-46. doi: 10.1002/cfg.381. Comp Funct Genomics. 2004. PMID: 18629061 Free PMC article.
-
Bacterial ammeline metabolism via guanine deaminase.J Bacteriol. 2010 Feb;192(4):1106-12. doi: 10.1128/JB.01243-09. Epub 2009 Dec 18. J Bacteriol. 2010. PMID: 20023034 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

