Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae
- PMID: 12200547
- PMCID: PMC129455
- DOI: 10.1073/pnas.182380799
Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae
Abstract
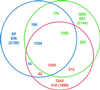
The 2,160,267 bp genome sequence of Streptococcus agalactiae, the leading cause of bacterial sepsis, pneumonia, and meningitis in neonates in the U.S. and Europe, is predicted to encode 2,175 genes. Genome comparisons among S. agalactiae, Streptococcus pneumoniae, Streptococcus pyogenes, and the other completely sequenced genomes identified genes specific to the streptococci and to S. agalactiae. These in silico analyses, combined with comparative genome hybridization experiments between the sequenced serotype V strain 2603 V/R and 19 S. agalactiae strains from several serotypes using whole-genome microarrays, revealed the genetic heterogeneity among S. agalactiae strains, even of the same serotype, and provided insights into the evolution of virulence mechanisms.
Figures


References
-
- Zangwill K. M., Schuchat, A. & Wenger, J. D. (1992) Mor. Mortal Wkly. Rep. CDC Surveill. Summ. 41, 25-32. - PubMed
-
- Schuchat A. & Wenger, J. D. (1994) Epidemiol. Rev. 16, 374-402. - PubMed
-
- Anonymous (1996) MMWR Recomm. Rep. 45, 1-24. - PubMed
-
- Harrison L. H., Elliott, J. A., Dwyer, D. M., Libonati, J. P., Ferrieri, P., Billmann, L. & Schuchat, A. (1998) J. Infect. Dis. 177, 998-1002. - PubMed
-
- Lin F. Y., Clemens, J. D., Azimi, P. H., Regan, J. A., Weisman, L. E., Philips, J. B., III, Rhoads, G. G., Clark, P., Brenner, R. A. & Ferrieri, P. (1998) J. Infect. Dis. 177, 790-792. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

