Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy
- PMID: 12202037
- PMCID: PMC4459650
- DOI: 10.1016/s0092-8674(02)00861-9
Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy
Abstract
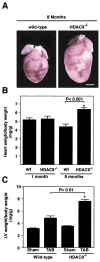
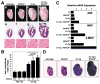
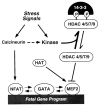
The heart responds to stress signals by hypertrophic growth, which is accompanied by activation of the MEF2 transcription factor and reprogramming of cardiac gene expression. We show here that class II histone deacetylases (HDACs), which repress MEF2 activity, are substrates for a stress-responsive kinase specific for conserved serines that regulate MEF2-HDAC interactions. Signal-resistant HDAC mutants lacking these phosphorylation sites are refractory to hypertrophic signaling and inhibit cardiomyocyte hypertrophy. Conversely, mutant mice lacking the class II HDAC, HDAC9, are sensitized to hypertrophic signals and exhibit stress-dependent cardiomegaly. Thus, class II HDACs act as signal-responsive suppressors of the transcriptional program governing cardiac hypertrophy and heart failure.
Figures

References
-
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. - PubMed
-
- Chien KR. Stress pathways and heart failure. Cell. 1999;98:555–558. - PubMed
-
- Frey N, McKinsey TA, Olson EN. Decoding calcium signals involved in cardiac growth and function. Nat Med. 2000;6:1221–1227. - PubMed
-
- Grozinger CM, Schreiber SL. Deacetylase enzymes. Biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9:3–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

