Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium
- PMID: 12202574
- PMCID: PMC130708
- DOI: 10.1128/JCM.40.9.3326-3333.2002
Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium
Abstract
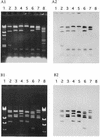
A total of 640 vancomycin-resistant Enterococcus faecium (VRE) isolates, which were obtained between 1994 and 1999 from the Medical School Hospital of the University of Michigan, Ann Arbor, were used in this study. Of the 640 strains, 611 and 29 were VanA and VanB VRE, respectively, based on PCR analysis. Four hundred ninety-two (77%) of the strains exhibited resistance to concentrations of gentamicin from 64 micro g/ml (MIC) to more than 1,024 micro g/ml (MIC). The gentamicin resistance of each of 261 (53%) of the 492 gentamicin-resistant strains was transferred to E. faecium at a frequency of about 10(-5) to 10(-6) per donor cell in broth mating. More than 90% of vancomycin resistances of the 261 strains cotransferred with the gentamicin resistances to E. faecium strains by filter mating. The conjugative gentamicin resistance plasmids were identified and were classified into five types (A through E) with respect to their EcoRI restriction profiles. The transfer frequencies of each type of plasmid between E. faecium strains or Enterococcus faecalis strains were around 10(-3) to 10(-5) per donor cell or around 10(-6) to 10(-7) per donor cell, respectively, in broth mating. Type A and type B were the most frequently isolated, at an isolation frequency of about 40% per VRE isolate harboring the gentamicin resistance conjugative plasmid. The plasmids did not show any homology in Southern hybridization with the pheromone-responsive plasmids and broad-host-range plasmids pAMbeta1 and pIP501. The EcoRI or NdeI restriction fragments of each type of plasmids hybridized to the conjugative gentamicin resistance plasmid pMG1 (65.1 kb), which was originally isolated from an E. faecium clinical isolate, and transfer efficiently in broth mating.
Figures

References
-
- Arthur, M., F. Depardieu, C. Molinas, P. Reynolds, P. Courvalin. 1995. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 154:87-92. - PubMed
-
- Behnke, D., and M. S. Gilmore. 1981. Location of antibiotic resistance determinants, copy control, and replication functions on the double-selective streptococcal cloning vector pGB301. Mol. Gen. Genet. 184:115-120. - PubMed
-
- Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

