Predicted structure and phyletic distribution of the RNA-binding protein Hfq
- PMID: 12202750
- PMCID: PMC137430
- DOI: 10.1093/nar/gkf508
Predicted structure and phyletic distribution of the RNA-binding protein Hfq
Abstract
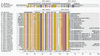
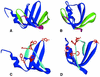
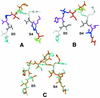
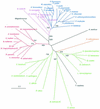
Hfq, a bacterial RNA-binding protein, was recently shown to contain the Sm1 motif, a characteristic of Sm and LSm proteins that function in RNA processing events in archaea and eukaryotes. In this report, comparative structural modeling was used to predict a three-dimensional structure of the Hfq core sequence. The predicted structure aligns with most major features of the Methanobacterium thermoautotrophicum LSm protein structure. Conserved residues in Hfq are positioned at the same structural locations responsible for subunit assembly and RNA interaction in Sm proteins. A highly conserved portion of Hfq assumes a structural fold similar to the Sm2 motif of Sm proteins. The evolution of the Hfq protein was explored by conducting a BLAST search of microbial genomes followed by phylogenetic analysis. Approximately half of the 140 complete or nearly complete genomes examined contain at least one gene coding for Hfq. The presence or absence of Hfq closely followed major bacterial clades. It is absent from high-level clades and present in the ancient Thermotogales-Aquificales clade and all proteobacteria except for those that have undergone major reduction in genome size. Residues at three positions in Hfq form signatures for the beta/gamma proteobacteria, alpha proteobacteria and low GC Gram-positive bacteria groups.
Figures


References
-
- Franze de Fernandez M.T., Eoyang,L. and August,J.T. (1968) Factor fraction required for the synthesis of bacteriophage Q beta-RNA. Nature, 219, 588–590. - PubMed
-
- Carmichael G.G., Weber,K., Niveleau,A. and Wahba,A.J. (1975) The host factor required for RNA phage Qbeta RNA replication in vitro. Intracellular location, quantitation, and purification by polyadenylate-cellulose chromatography. J. Biol. Chem., 250, 3607–3612. - PubMed
-
- Azam T.A, Hiraga,S. and Ishihama,A. (2000) Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells, 5, 613–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

