Mutations altering the cleavage specificity of a homing endonuclease
- PMID: 12202772
- PMCID: PMC137417
- DOI: 10.1093/nar/gkf495
Mutations altering the cleavage specificity of a homing endonuclease
Abstract
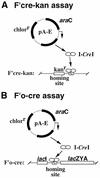
The homing endonuclease I-CreI recognizes and cleaves a particular 22 bp DNA sequence. The crystal structure of I-CreI bound to homing site DNA has previously been determined, leading to a number of predictions about specific protein-DNA contacts. We test these predictions by analyzing a set of endonuclease mutants and a complementary set of homing site mutants. We find evidence that all structurally predicted I-CreI/DNA contacts contribute to DNA recognition and show that these contacts differ greatly in terms of their relative importance. We also describe the isolation of a collection of altered specificity I-CreI derivatives. The in vitro DNA-binding and cleavage properties of two such endonucleases demonstrate that our genetic approach is effective in identifying homing endonucleases that recognize and cleave novel target sequences.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

