Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells
- PMID: 12208843
- PMCID: PMC186665
- DOI: 10.1101/gad.230702
Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells
Abstract
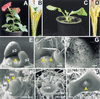
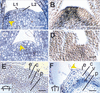
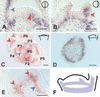
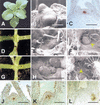
Plant shoot development depends on the perpetuation of a group of undifferentiated cells in the shoot apical meristem (SAM). In the Petunia mutant hairy meristem (ham), shoot meristems differentiate postembryonically as continuations of the subtending stem. HAM encodes a putative transcription factor of the GRAS family, which acts non-cell-autonomously from L3-derived tissue of lateral organ primordia and stem provasculature. HAM acts in parallel with TERMINATOR (PhWUSCHEL) and is required for continued cellular response to TERMINATOR and SHOOTMERISTEMLESS (PhSTM). This reveals a novel mechanism by which signals from differentiating tissues extrinsically control stem cell fate in the shoot apex.
Figures

References
-
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsison a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. - PubMed
-
- Byrne M, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. - PubMed
-
- Janssen BJ, Williams A, Chen JJ, Mathern J, Hake S, Sinha N. Isolation and characterisation of two knotted-like homeobox genes from tomato. Plant Mol Biol. 1998;36:417–425. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
