Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing
- PMID: 12208851
- PMCID: PMC186666
- DOI: 10.1101/gad.991102
Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing
Abstract
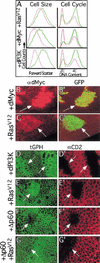
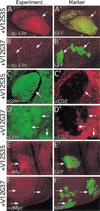
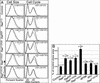
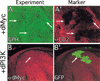
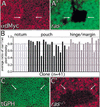
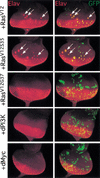
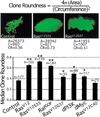
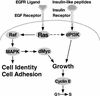
The Ras GTPase links extracellular signals to intracellular mechanisms that control cell growth, the cell cycle, and cell identity. An activated form of Drosophila Ras (Ras(V12)) promotes these processes in the developing wing, but the effector pathways involved are unclear. Here, we present evidence indicating that Ras(V12) promotes cell growth and G(1)/S progression by increasing dMyc protein levels and activating dPI3K signaling, and that it does so via separate effector pathways. We also show that endogenous Ras is required to maintain normal levels of dMyc, but not dPI3K signaling during wing development. Finally, we show that induction of dMyc and regulation of cell identity are separable effects of Raf/MAPK signaling. These results suggest that Ras may only affect PI3K signaling when mutationally activated, such as in Ras(V12)-transformed cells, and provide a basis for understanding the synergy between Ras and other growth-promoting oncogenes in cancer.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
