Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase
- PMID: 12208995
- PMCID: PMC120795
- DOI: 10.1128/MMBR.66.3.373-395.2002
Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase
Abstract
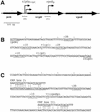
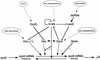
The sigma(S) (RpoS) subunit of RNA polymerase is the master regulator of the general stress response in Escherichia coli and related bacteria. While rapidly growing cells contain very little sigma(S), exposure to many different stress conditions results in rapid and strong sigma(S) induction. Consequently, transcription of numerous sigma(S)-dependent genes is activated, many of which encode gene products with stress-protective functions. Multiple signal integration in the control of the cellular sigma(S) level is achieved by rpoS transcriptional and translational control as well as by regulated sigma(S) proteolysis, with various stress conditions differentially affecting these levels of sigma(S) control. Thus, a reduced growth rate results in increased rpoS transcription whereas high osmolarity, low temperature, acidic pH, and some late-log-phase signals stimulate the translation of already present rpoS mRNA. In addition, carbon starvation, high osmolarity, acidic pH, and high temperature result in stabilization of sigma(S), which, under nonstress conditions, is degraded with a half-life of one to several minutes. Important cis-regulatory determinants as well as trans-acting regulatory factors involved at all levels of sigma(S) regulation have been identified. rpoS translation is controlled by several proteins (Hfq and HU) and small regulatory RNAs that probably affect the secondary structure of rpoS mRNA. For sigma(S) proteolysis, the response regulator RssB is essential. RssB is a specific direct sigma(S) recognition factor, whose affinity for sigma(S) is modulated by phosphorylation of its receiver domain. RssB delivers sigma(S) to the ClpXP protease, where sigma(S) is unfolded and completely degraded. This review summarizes our current knowledge about the molecular functions and interactions of these components and tries to establish a framework for further research on the mode of multiple signal input into this complex regulatory system.
Figures
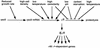

References
-
- Altuvia, S., D. Weinstein-Fischer, A. Zhang, L. Postow, and G. Storz. 1997. A small, stable RNA induced by oxidative stress: roles as a pleiotropic regulator and antimutator. Cell 90:43-53. - PubMed
-
- Andersson, R. A., E. T. Palva, and M. Pirhonen. 1999. The response regulator ExpM is essential for the virulence of Erwinia carotovora subsp. carotovora and acts negatively on the sigma factor RpoS (σS). Mol. Plant-Microbe Interact. 12:575-584. - PubMed
-
- Argaman, L., and S. Altuvia. 2000. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 300:1101-1112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

