ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms
- PMID: 12215510
- PMCID: PMC150760
- DOI: 10.1105/tpc.003772
ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms
Abstract
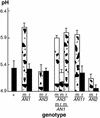
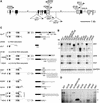
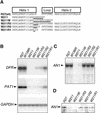
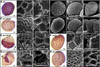
ANTHOCYANIN1 (AN1) of petunia is a transcription factor of the basic helix-loop-helix (bHLH) family that is required for the synthesis of anthocyanin pigments. Here, we show that AN1 controls additional aspects of cell differentiation: the acidification of vacuoles in petal cells, and the size and morphology of cells in the seed coat epidermis. We identified an1 alleles, formerly known as ph6, that sustain anthocyanin synthesis but not vacuolar acidification and seed coat morphogenesis. These alleles express truncated proteins lacking the C-terminal half of AN1, including the bHLH domain, at an approximately 30-fold higher level than wild-type AN1. An allelic series in which one, two, or three amino acids were inserted into the bHLH domain indicated that this domain is required for both anthocyanin synthesis and vacuolar acidification. These findings show that AN1 controls more aspects of epidermal cell differentiation than previously thought through partially separable domains.
Figures


Similar articles
-
PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway.Plant Cell. 2006 May;18(5):1274-91. doi: 10.1105/tpc.105.034041. Epub 2006 Apr 7. Plant Cell. 2006. PMID: 16603655 Free PMC article.
-
anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes.Plant Cell. 2000 Sep;12(9):1619-32. doi: 10.1105/tpc.12.9.1619. Plant Cell. 2000. PMID: 11006336 Free PMC article.
-
Functionally Similar WRKY Proteins Regulate Vacuolar Acidification in Petunia and Hair Development in Arabidopsis.Plant Cell. 2016 Mar;28(3):786-803. doi: 10.1105/tpc.15.00608. Epub 2016 Mar 14. Plant Cell. 2016. PMID: 26977085 Free PMC article.
-
A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation.Plant J. 2007 Feb;49(4):641-54. doi: 10.1111/j.1365-313X.2006.02988.x. Epub 2007 Jan 18. Plant J. 2007. PMID: 17270013
-
Proteomics of red and white corolla limbs in petunia reveals a novel function of the anthocyanin regulator ANTHOCYANIN1 in determining flower longevity.J Proteomics. 2016 Jan 10;131:38-47. doi: 10.1016/j.jprot.2015.10.008. Epub 2015 Oct 13. J Proteomics. 2016. PMID: 26459403
Cited by
-
Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1.Sci Rep. 2016 Sep 1;6:32534. doi: 10.1038/srep32534. Sci Rep. 2016. PMID: 27581206 Free PMC article.
-
Differential timing of spider mite-induced direct and indirect defenses in tomato plants.Plant Physiol. 2004 May;135(1):483-95. doi: 10.1104/pp.103.038315. Epub 2004 Apr 30. Plant Physiol. 2004. PMID: 15122016 Free PMC article.
-
Evolutionary comparison of competitive protein-complex formation of MYB, bHLH, and WDR proteins in plants.J Exp Bot. 2019 Jun 28;70(12):3197-3209. doi: 10.1093/jxb/erz155. J Exp Bot. 2019. PMID: 31071215 Free PMC article.
-
Discrete bHLH transcription factors play functionally overlapping roles in pigmentation patterning in flowers of Antirrhinum majus.New Phytol. 2021 Jul;231(2):849-863. doi: 10.1111/nph.17142. Epub 2021 Jan 12. New Phytol. 2021. PMID: 33616943 Free PMC article.
-
Flavonoids - flowers, fruit, forage and the future.J R Soc N Z. 2022 Feb 28;53(3):304-331. doi: 10.1080/03036758.2022.2034654. eCollection 2023. J R Soc N Z. 2022. PMID: 39439482 Free PMC article. Review.
References
-
- Bianchi, F., Cornelissen, P.J.T., Gerats, A.G.M., and Hogervorst, J.M.W. (1978). Regulation of gene action in Petunia hybrida: Unstable alleles of a gene for flower colour. Theor. Appl. Genet. 53, 157–167. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

