Arabidopsis A BOUT DE SOUFFLE, which is homologous with mammalian carnitine acyl carrier, is required for postembryonic growth in the light
- PMID: 12215513
- PMCID: PMC150763
- DOI: 10.1105/tpc.002485
Arabidopsis A BOUT DE SOUFFLE, which is homologous with mammalian carnitine acyl carrier, is required for postembryonic growth in the light
Abstract
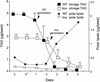
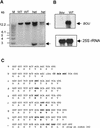
The degradation of storage compounds just after germination is essential to plant development, providing energy and molecules necessary for the building of a photosynthetic apparatus and allowing autotrophic growth. We identified à bout de souffle (bou), a new Arabidopsis mutation. Mutant plants stopped developing after germination and degraded storage lipids, but they did not proceed to autotrophic growth. Neither leaves nor roots developed in the mutant. However, externally added sugar or germination in the dark could bypass this developmental block and allowed mutant plants to develop. The mutated gene was cloned using the transposon Dissociation as a molecular tag. The gene coding sequence showed similarity to those of the mitochondrial carnitine acyl carriers (CACs) or CAC-like proteins. In animals and yeast, these transmembrane proteins are involved in the transport of lipid-derived molecules across mitochondrial membranes for energy and carbon supply. The data presented here suggest that BOU identifies a novel mitochondrial pathway that is necessary to seedling development in the light. The BOU pathway would be an alternative to the well-known glyoxylate pathway.
Figures

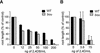




Similar articles
-
Arabidopsis A BOUT DE SOUFFLE is a putative mitochondrial transporter involved in photorespiratory metabolism and is required for meristem growth at ambient CO₂ levels.Plant J. 2013 Mar;73(5):836-49. doi: 10.1111/tpj.12082. Epub 2013 Feb 12. Plant J. 2013. PMID: 23181524
-
The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein alpha-subunit GPA1 and regulates seed germination and early seedling development.Plant Cell. 2003 Jul;15(7):1578-90. doi: 10.1105/tpc.011890. Plant Cell. 2003. PMID: 12837948 Free PMC article.
-
A novel zinc-finger protein with a proline-rich domain mediates ABA-regulated seed dormancy in Arabidopsis.Plant Mol Biol. 2004 Jan;54(1):1-9. doi: 10.1023/B:PLAN.0000028730.10834.e3. Plant Mol Biol. 2004. PMID: 15159630
-
A nuclear-encoded mitochondrial gene AtCIB22 is essential for plant development in Arabidopsis.J Genet Genomics. 2010 Oct;37(10):667-83. doi: 10.1016/S1673-8527(09)60085-0. J Genet Genomics. 2010. PMID: 21035093
-
AtDSEL, an Arabidopsis cytosolic DAD1-like acylhydrolase, is involved in negative regulation of storage oil mobilization during seedling establishment.J Plant Physiol. 2011 Sep 15;168(14):1705-9. doi: 10.1016/j.jplph.2011.03.004. Epub 2011 Apr 8. J Plant Physiol. 2011. PMID: 21477884
Cited by
-
Proteomic identification and characterization of a novel peroxisomal adenine nucleotide transporter supplying ATP for fatty acid beta-oxidation in soybean and Arabidopsis.Plant Cell. 2008 Dec;20(12):3227-40. doi: 10.1105/tpc.108.062877. Epub 2008 Dec 10. Plant Cell. 2008. PMID: 19073762 Free PMC article.
-
CIPK9 is involved in seed oil regulation in Brassica napus L. and Arabidopsis thaliana (L.) Heynh.Biotechnol Biofuels. 2018 May 2;11:124. doi: 10.1186/s13068-018-1122-z. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29743952 Free PMC article.
-
Arabidopsis seedlings deficient in a plastidic pyruvate kinase are unable to utilize seed storage compounds for germination and establishment.Plant Physiol. 2007 Dec;145(4):1670-80. doi: 10.1104/pp.107.108340. Epub 2007 Oct 26. Plant Physiol. 2007. PMID: 17965177 Free PMC article.
-
Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins.Plant Cell. 2004 Jan;16(1):241-56. doi: 10.1105/tpc.016055. Epub 2003 Dec 11. Plant Cell. 2004. PMID: 14671022 Free PMC article.
-
The plant mitochondrial carrier family: functional and evolutionary aspects.Front Plant Sci. 2012 Jan 18;3:2. doi: 10.3389/fpls.2012.00002. eCollection 2012. Front Plant Sci. 2012. PMID: 22639632 Free PMC article.
References
-
- ap Rees, T. (1987). Compartmentation of plant metabolism. In The Biochemistry of Plants: A Comprehensive Treatise, Vol. 12, P.K. Stumpf and E.E. Conn, eds (New York: Academic Press), pp. 87–115.
-
- Bao, X., Focke, M., Pollard, M., and Ohlrogge, J. (2000). Understanding in vivo carbon precursor for fatty acid synthesis in leaf tissue. Plant J. 22, 39–50. - PubMed
-
- Carol, P., Stevenson, D., Bisanz, C., Breitenbach, J., Sandmann, G., Coupland, G., Mache, R., and Kuntz, M. (1999). Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11, 57–68. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

