Loss of Albino3 leads to the specific depletion of the light-harvesting system
- PMID: 12215522
- PMCID: PMC150772
- DOI: 10.1105/tpc.003442
Loss of Albino3 leads to the specific depletion of the light-harvesting system
Abstract
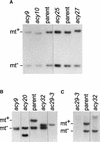
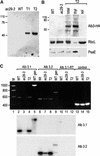
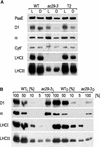
The chloroplast Albino3 (Alb3) protein is a chloroplast homolog of the mitochondrial Oxa1p and YidC proteins of Escherichia coli, which are essential components for integrating membrane proteins. In vitro studies in vascular plants have revealed that Alb3 is required for the integration of the light-harvesting complex protein into the thylakoid membrane. Here, we show that the gene affected in the ac29 mutant of Chlamydomonas reinhardtii is Alb3.1. The availability of the ac29 mutant has allowed us to examine the function of Alb3.1 in vivo. The loss of Alb3.1 has two major effects. First, the amount of light-harvesting complex from photosystem II (LHCII) and photosystem I (LHCI) is reduced >10-fold, and total chlorophyll represents only 30% of wild-type levels. Second, the amount of photosystem II is diminished 2-fold in light-grown cells and nearly 10-fold in dark-grown cells. The accumulation of photosystem I, the cytochrome b(6)f complex, and ATP synthase is not affected in the ac29 mutant. Mild solubilization of thylakoid membranes reveals that Alb3 forms two distinct complexes, a lower molecular mass complex of a size similar to LHC and a high molecular mass complex. A homolog of Alb3.1, Alb3.2, is present in Chlamydomonas, with 37% sequence identity and 57% sequence similarity. Based on the phenotype of ac29, these two genes appear to have mostly nonredundant functions.
Figures




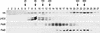
Similar articles
-
Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3.Plant Cell. 2004 Jul;16(7):1790-800. doi: 10.1105/tpc.023226. Epub 2004 Jun 18. Plant Cell. 2004. PMID: 15208384 Free PMC article.
-
Assembly Apparatus of Light-Harvesting Complexes: Identification of Alb3.1-cpSRP-LHCP Complexes in the Green Alga Chlamydomonas reinhardtii.Plant Cell Physiol. 2022 Jan 25;63(1):70-81. doi: 10.1093/pcp/pcab146. Plant Cell Physiol. 2022. PMID: 34592750
-
The light-harvesting complex of photosystem I in Chlamydomonas reinhardtii: protein composition, gene structures and phylogenic implications.Plant Cell Physiol. 2004 Feb;45(2):138-45. doi: 10.1093/pcp/pch013. Plant Cell Physiol. 2004. PMID: 14988484
-
A genome's-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii.Curr Genet. 2004 Feb;45(2):61-75. doi: 10.1007/s00294-003-0460-x. Epub 2003 Dec 2. Curr Genet. 2004. PMID: 14652691 Review.
-
Structural analysis and comparison of light-harvesting complexes I and II.Biochim Biophys Acta Bioenerg. 2020 Apr 1;1861(4):148038. doi: 10.1016/j.bbabio.2019.06.010. Epub 2019 Jun 20. Biochim Biophys Acta Bioenerg. 2020. PMID: 31229568 Review.
Cited by
-
Functional Update of the Auxiliary Proteins PsbW, PsbY, HCF136, PsbN, TerC and ALB3 in Maintenance and Assembly of PSII.Front Plant Sci. 2016 Apr 7;7:423. doi: 10.3389/fpls.2016.00423. eCollection 2016. Front Plant Sci. 2016. PMID: 27092151 Free PMC article. Review.
-
LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana.Plant Cell. 2007 Jun;19(6):1980-93. doi: 10.1105/tpc.107.050526. Epub 2007 Jun 29. Plant Cell. 2007. Retraction in: Plant Cell. 2016 Dec;28(12):3061. doi: 10.1105/tpc.16.00881. PMID: 17601825 Free PMC article. Retracted.
-
Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3.Plant Cell. 2004 Jul;16(7):1790-800. doi: 10.1105/tpc.023226. Epub 2004 Jun 18. Plant Cell. 2004. PMID: 15208384 Free PMC article.
-
The terminal enzymes of (bacterio)chlorophyll biosynthesis.R Soc Open Sci. 2022 May 4;9(5):211903. doi: 10.1098/rsos.211903. eCollection 2022 May. R Soc Open Sci. 2022. PMID: 35573041 Free PMC article. Review.
-
One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas.Plant Cell. 2006 Jun;18(6):1454-66. doi: 10.1105/tpc.105.038695. Epub 2006 May 5. Plant Cell. 2006. PMID: 16679460 Free PMC article.
References
-
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. - PubMed
-
- Bartsevich, V.V., and Pakrasi, H.B. (1997). Molecular identification of a novel protein that regulates biogenesis of photosystem I, a membrane protein complex. J. Biol. Chem. 272, 6382–6387. - PubMed
-
- Bonnefoy, N., Chalvet, F., Hamel, P., Slonimski, P.P., and Dujardin, G. (1994). OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes, controls cytochrome oxidase biogenesis. J. Mol. Biol. 239, 201–212. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

