NRC-interacting factor 1 is a novel cotransducer that interacts with and regulates the activity of the nuclear hormone receptor coactivator NRC
- PMID: 12215545
- PMCID: PMC134037
- DOI: 10.1128/MCB.22.19.6883-6894.2002
NRC-interacting factor 1 is a novel cotransducer that interacts with and regulates the activity of the nuclear hormone receptor coactivator NRC
Abstract
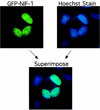
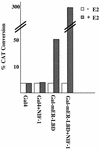
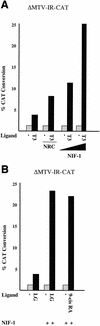
We previously reported the cloning and characterization of a novel nuclear hormone receptor transcriptional coactivator, which we refer to as NRC. NRC is a 2,063-amino-acid nuclear protein which contains a potent N-terminal activation domain and several C-terminal modules which interact with CBP and ligand-bound nuclear hormone receptors as well as c-Fos and c-Jun. In this study we sought to clone and identify novel factors that interact with NRC to modulate its transcriptional activity. Here we describe the cloning and characterization of a novel protein we refer to as NIF-1 (NRC-interacting factor 1). NIF-1 was cloned from rat pituitary and human cell lines and was found to interact in vivo and in vitro with NRC. NIF-1 is a 1,342-amino-acid nuclear protein containing a number of conserved domains, including six Cys-2/His-2 zinc fingers, an N-terminal stretch of acidic amino acids, and a C-terminal leucine zipper-like motif. Zinc fingers 1 to 3 are potential DNA-binding BED finger domains recently proposed to play a role in altering local chromatin architecture. We mapped the interaction domains of NRC and NIF-1. Although NIF-1 does not directly interact with nuclear receptors, it markedly enhances ligand-dependent transcriptional activation by nuclear hormone receptors in vivo as well as activation by c-Fos and c-Jun. These results, and the finding that NIF-1 interacts with NRC in vivo, suggest that NIF-1 functions to regulate transcriptional activation through NRC. We suggest that NIF-1, and factors which associate with coactivators but not receptors, be referred to as cotransducers, which act in vivo either as part of a coactivator complex or downstream of a coactivator complex to modulate transcriptional activity. Our findings suggest that NIF-1 may be a functional component of an NRC complex and acts as a regulator or cotransducer of NRC function.
Figures







References
-
- Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. - PubMed
-
- Aranda, A., and A. Pascual. 2001. Nuclear hormone receptors and gene expression. Physiol. Rev. 81:1269-1304. - PubMed
-
- Aravind, L. 2000. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem. Sci. 25:421-423. - PubMed
-
- Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
