A new modified ortho cleavage pathway of 3-chlorocatechol degradation by Rhodococcus opacus 1CP: genetic and biochemical evidence
- PMID: 12218013
- PMCID: PMC135353
- DOI: 10.1128/JB.184.19.5282-5292.2002
A new modified ortho cleavage pathway of 3-chlorocatechol degradation by Rhodococcus opacus 1CP: genetic and biochemical evidence
Abstract
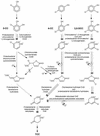
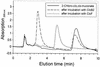
The 4-chloro- and 2,4-dichlorophenol-degrading strain Rhodococcus opacus 1CP has previously been shown to acquire, during prolonged adaptation, the ability to mineralize 2-chlorophenol. In addition, homogeneous chlorocatechol 1,2-dioxygenase from 2-chlorophenol-grown biomass has shown relatively high activity towards 3-chlorocatechol. Based on sequences of the N terminus and tryptic peptides of this enzyme, degenerate PCR primers were now designed and used for cloning of the respective gene from genomic DNA of strain 1CP. A 9.5-kb fragment containing nine open reading frames was obtained on pROP1. Besides other genes, a gene cluster consisting of four chlorocatechol catabolic genes was identified. As judged by sequence similarity and correspondence of predicted N termini with those of purified enzymes, the open reading frames correspond to genes for a second chlorocatechol 1,2-dioxygenase (ClcA2), a second chloromuconate cycloisomerase (ClcB2), a second dienelactone hydrolase (ClcD2), and a muconolactone isomerase-related enzyme (ClcF). All enzymes of this new cluster are only distantly related to the known chlorocatechol enzymes and appear to represent new evolutionary lines of these activities. UV overlay spectra as well as high-pressure liquid chromatography analyses confirmed that 2-chloro-cis,cis-muconate is transformed by ClcB2 to 5-chloromuconolactone, which during turnover by ClcF gives cis-dienelactone as the sole product. cis-Dienelactone was further hydrolyzed by ClcD2 to maleylacetate. ClcF, despite its sequence similarity to muconolactone isomerases, no longer showed muconolactone-isomerizing activity and thus represents an enzyme dedicated to its new function as a 5-chloromuconolactone dehalogenase. Thus, during 3-chlorocatechol degradation by R. opacus 1CP, dechlorination is catalyzed by a muconolactone isomerase-related enzyme rather than by a specialized chloromuconate cycloisomerase.
Figures



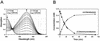
Similar articles
-
Conversion of 2-fluoromuconate to cis-dienelactone by purified enzymes of Rhodococcus opacus 1cp.Appl Environ Microbiol. 2003 Sep;69(9):5636-42. doi: 10.1128/AEM.69.9.5636-5642.2003. Appl Environ Microbiol. 2003. PMID: 12957954 Free PMC article.
-
Evolutionary relationship between chlorocatechol catabolic enzymes from Rhodococcus opacus 1CP and their counterparts in proteobacteria: sequence divergence and functional convergence.J Bacteriol. 1998 Mar;180(5):1082-94. doi: 10.1128/JB.180.5.1082-1094.1998. J Bacteriol. 1998. PMID: 9495745 Free PMC article.
-
Enzymes of a new modified ortho-pathway utilizing 2-chlorophenol in Rhodococcus opacus 1CP.Biochemistry (Mosc). 2001 May;66(5):548-55. doi: 10.1023/a:1010267104238. Biochemistry (Mosc). 2001. PMID: 11405892
-
Evolution of chlorocatechol catabolic pathways. Conclusions to be drawn from comparisons of lactone hydrolases.Biodegradation. 1994 Dec;5(3-4):301-21. doi: 10.1007/BF00696467. Biodegradation. 1994. PMID: 7765840 Review.
-
Biochemical features of the degradation of pollutants by Rhodococcus as a basis for contaminated wastewater and soil cleanup.Mikrobiologiia. 2011 Sep-Oct;80(5):579-94. Mikrobiologiia. 2011. PMID: 22168001 Review.
Cited by
-
Conversion of 2-fluoromuconate to cis-dienelactone by purified enzymes of Rhodococcus opacus 1cp.Appl Environ Microbiol. 2003 Sep;69(9):5636-42. doi: 10.1128/AEM.69.9.5636-5642.2003. Appl Environ Microbiol. 2003. PMID: 12957954 Free PMC article.
-
Bacterial degradation of chlorophenols and their derivatives.Microb Cell Fact. 2014 Mar 3;13(1):31. doi: 10.1186/1475-2859-13-31. Microb Cell Fact. 2014. PMID: 24589366 Free PMC article. Review.
-
Kinetics of interaction between substrates/substrate analogs and benzoate 1,2-dioxygenase from benzoate-degrading Rhodococcus opacus 1CP.Folia Microbiol (Praha). 2017 Jul;62(4):355-362. doi: 10.1007/s12223-017-0505-z. Epub 2017 Feb 24. Folia Microbiol (Praha). 2017. PMID: 28236176
-
Novel acetone metabolism in a propane-utilizing bacterium, Gordonia sp. strain TY-5.J Bacteriol. 2007 Feb;189(3):886-93. doi: 10.1128/JB.01054-06. Epub 2006 Oct 27. J Bacteriol. 2007. PMID: 17071761 Free PMC article.
-
Unveiling novel features and phylogenomic assessment of indigenous Priestia megaterium AB-S79 using comparative genomics.Microbiol Spectr. 2025 Apr;13(4):e0146624. doi: 10.1128/spectrum.01466-24. Epub 2025 Feb 19. Microbiol Spectr. 2025. PMID: 39969228 Free PMC article.
References
-
- Blasco, R., R. M. Wittich, M. Mallavarapu, K. N. Timmis, and D. H. Pieper. 1995. From xenobiotic to antibiotic, formation of protoanemonin from 4-chlorocatechol by enzymes of the 3-oxoadipate pathway. J. Biol. Chem. 270:29229-29235. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, S. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, S. Skelton, S. Squares, R. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. - PubMed
-
- Dorn, E., M. Hellwig, W. Reineke, and H.-J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

