Interdependent expression of the ccoNOQP-rdxBHIS loci in Rhodobacter sphaeroides 2.4.1
- PMID: 12218019
- PMCID: PMC135364
- DOI: 10.1128/JB.184.19.5330-5338.2002
Interdependent expression of the ccoNOQP-rdxBHIS loci in Rhodobacter sphaeroides 2.4.1
Abstract
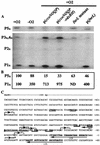
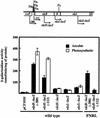
The rdxBHIS gene cluster of Rhodobacter sphaeroides 2.4.1, located downstream of the ccoNOQP operon encoding the cbb(3) cytochrome c oxidase, is required for the posttranscriptional modification of the cbb(3) cytochrome c oxidase. The cbb(3) cytochrome c oxidase is the main terminal oxidase under microaerobic conditions, as well as a component of the signal transduction pathway controlling photosynthesis gene expression. Because of the intimate functional and positional relationships of the ccoNOQP operon and the rdxBHIS gene cluster, we have examined the transcriptional activities of this DNA region in order to understand their expression and regulation. Northern blot analysis and reverse transcription-PCR, together with earlier complementation analysis, suggested that the ccoNOQP-rdxBHIS cluster is transcribed as ccoNOQP-, ccoNOQP-rdxBH-, rdxBH-, and rdxIS-specific transcripts. Multiple transcriptional start sites have been identified by primer extension analyses: five for ccoN, four for rdxB, and one for rdxI. Transcription from P1(N) of ccoN and P1(B) of rdxB is dependent on the presence of FnrL. LacZ fusion analysis support the above-described studies, especially the importance of FnrL. Expression of the cco-rdx cluster is closely related to photosynthesis gene expression, suggesting that transcript stoichiometry and presumably the stoichiometry of the gene products are critical factors in controlling photosynthesis gene expression.
Figures





Similar articles
-
Evidence for the role of redox carriers in photosynthesis gene expression and carotenoid biosynthesis in Rhodobacter sphaeroides 2.4.1.J Bacteriol. 1997 Mar;179(6):1951-61. doi: 10.1128/jb.179.6.1951-1961.1997. J Bacteriol. 1997. PMID: 9068641 Free PMC article.
-
Genetic and phenotypic analyses of the rdx locus of Rhodobacter sphaeroides 2.4.1.J Bacteriol. 2000 Jun;182(12):3475-81. doi: 10.1128/JB.182.12.3475-3481.2000. J Bacteriol. 2000. PMID: 10852880 Free PMC article.
-
From redox flow to gene regulation: role of the PrrC protein of Rhodobacter sphaeroides 2.4.1.Biochemistry. 2000 Feb 29;39(8):2052-62. doi: 10.1021/bi9923858. Biochemistry. 2000. PMID: 10684655
-
Generalized approach to the regulation and integration of gene expression.Mol Microbiol. 2001 Mar;39(5):1116-23. doi: 10.1111/j.1365-2958.2001.02299.x. Mol Microbiol. 2001. PMID: 11251830 Review.
-
Regulation of photosynthetic gene expression in purple bacteria.Microbiology (Reading). 1998 Feb;144 ( Pt 2):267-278. doi: 10.1099/00221287-144-2-267. Microbiology (Reading). 1998. PMID: 9493364 Review. No abstract available.
Cited by
-
The use of chromatin immunoprecipitation to define PpsR binding activity in Rhodobacter sphaeroides 2.4.1.J Bacteriol. 2008 Oct;190(20):6817-28. doi: 10.1128/JB.00719-08. Epub 2008 Aug 8. J Bacteriol. 2008. PMID: 18689484 Free PMC article.
-
Functional domains of NosR, a novel transmembrane iron-sulfur flavoprotein necessary for nitrous oxide respiration.J Bacteriol. 2005 Mar;187(6):1992-2001. doi: 10.1128/JB.187.6.1992-2001.2005. J Bacteriol. 2005. PMID: 15743947 Free PMC article.
-
Transcriptional response of the photoheterotrophic marine bacterium Dinoroseobacter shibae to changing light regimes.ISME J. 2011 Dec;5(12):1957-68. doi: 10.1038/ismej.2011.68. Epub 2011 Jun 9. ISME J. 2011. PMID: 21654848 Free PMC article.
-
Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes.J Bacteriol. 2004 Jul;186(14):4748-58. doi: 10.1128/JB.186.14.4748-4758.2004. J Bacteriol. 2004. PMID: 15231807 Free PMC article.
-
Transcriptome and physiological responses to hydrogen peroxide of the facultatively phototrophic bacterium Rhodobacter sphaeroides.J Bacteriol. 2005 Nov;187(21):7232-42. doi: 10.1128/JB.187.21.7232-7242.2005. J Bacteriol. 2005. PMID: 16237007 Free PMC article.
References
-
- Eraso, J. M., and S. Kaplan. 2000. From redox flow to gene regulation: role of the PrrC protein of Rhodobacter sphaeroides 2.4.1. Biochemistry 39:2052-2062. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

