Forespore signaling is necessary for pro-sigmaK processing during Bacillus subtilis sporulation despite the loss of SpoIVFA upon translational arrest
- PMID: 12218026
- PMCID: PMC135367
- DOI: 10.1128/JB.184.19.5393-5401.2002
Forespore signaling is necessary for pro-sigmaK processing during Bacillus subtilis sporulation despite the loss of SpoIVFA upon translational arrest
Abstract
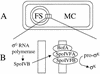
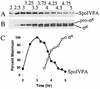
The sigmaK checkpoint coordinates gene expression in the mother cell with signaling from the forespore during Bacillus subtilis sporulation. The signaling pathway involves SpoIVB, a serine peptidase produced in the forespore, which is believed to cross the innermost membrane surrounding the forespore and activate a complex of proteins, including BofA, SpoIVFA, and SpoIVFB, located in the outermost membrane surrounding the forespore. Activation of the complex allows proteolytic processing of pro-sigmaK, and the resulting sigmaK RNA polymerase transcribes genes in the mother cell. To investigate activation of the pro-sigmaK processing complex, the level of SpoIVFA in extracts of sporulating cells was examined by Western blot analysis. The SpoIVFA level decreased when pro-sigmaK processing began during sporulation. In extracts of a spoIVB mutant defective in forespore signaling, the SpoIVFA level failed to decrease normally and no processing of pro-sigmaK was observed. Although these results are consistent with a model in which SpoIVFA inhibits processing until the SpoIVB-mediated signal is received from the forespore, we discovered that loss of SpoIVFA was insufficient to allow processing under certain conditions, including static incubation of the culture and continued shaking after the addition of inhibitors of oxidative phosphorylation or translation. Under these conditions, loss of SpoIVFA was independent of spoIVB. The inability to process pro-sigmaK under these conditions was not due to loss of SpoIVFB, the putative processing enzyme, or to a requirement for ongoing synthesis of pro-sigmaK. Rather, it was found that the requirements for shaking of the culture, for oxidative phosphorylation, and for translation could be bypassed by mutations that uncouple processing from dependence on forespore signaling. This suggests that ongoing translation is normally required for efficient pro-sigmaK processing because synthesis of the SpoIVB signal protein is needed to activate the processing complex. When translation is blocked, synthesis of SpoIVB ceases, and the processing complex remains inactive despite the loss of SpoIVFA. Taken together, the results suggest that SpoIVB signaling activates the processing complex by performing another function in addition to causing loss of SpoIVFA or by causing loss of SpoIVFA in a different way than when translation is blocked. The results also demonstrate that the processing machinery can function in the absence of translation or an electrochemical gradient across membranes.
Figures







References
-
- Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. - PubMed
-
- Cutting, S., A. Driks, R. Schmidt, B. Kunkel, and R. Losick. 1991. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-σK processing in Bacillus subtilis. Genes Dev. 5:456-466. - PubMed
-
- Cutting, S., V. Oke, A. Driks, R. Losick, S. Lu, and L. Kroos. 1990. A forespore checkpoint for mother-cell gene expression during development in Bacillus subtilis. Cell 62:239-250. - PubMed
-
- Cutting, S., S. Roels, and R. Losick. 1991. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J. Mol. Biol. 221:1237-1256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

