sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4
- PMID: 12218034
- PMCID: PMC135357
- DOI: 10.1128/JB.184.19.5457-5467.2002
sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4
Abstract
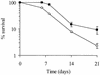
The accessory sigma factor sigmaB controls a general stress response that is thought to be important for Staphylococcus aureus survival and may contribute to virulence. The strain of choice for genetic studies, 8325-4, carries a small deletion in rsbU, which encodes a positive regulator of sigmaB activity. Consequently, to enable the role of sigmaB in virulence to be addressed, we constructed an rsbU(+) derivative, SH1000, using a method that does not leave behind an antibiotic resistance marker. The phenotypic properties of SH1000 (8325-4 rsbU(+)) were characterized and compared to those of 8325-4, the rsbU mutant, parent strain. A recognition site for sigmaB was located in the promoter region of katA, the gene encoding the sole catalase of S. aureus, by primer extension analysis. However, catalase expression and activity were similar in SH1000 (8325-4 rsbU(+)), suggesting that this promoter may have a minor role in catalase expression under normal conditions. Restoration of sigmaB activity in SH1000 (8325-4 rsbU(+)) resulted in a marked decrease in the levels of the exoproteins SspA and Hla, and this is likely to be mediated by reduced expression of agr in this strain. By using Western blotting and a sarA-lacZ reporter assay, the levels of SarA were found to be similar in strains 8325-4 and SH1000 (8325-4 rsbU(+)) and sigB mutant derivatives of these strains. This finding contrasts with previous reports that suggested that SarA expression levels are altered when they are measured transcriptionally. Inactivation of sarA in each of these strains resulted in an expected decrease in agr expression; however, the relative level of agr in SH1000 (8325-4 rsbU(+)) remained less than the relative levels in 8325-4 and the sigB mutant derivatives. We suggest that SarA is not likely to be the effector in the overall sigmaB-mediated effect on agr expression.
Figures


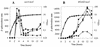

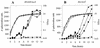

References
-
- Beers, R. F., and I. W. Sizer. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195:133-140. - PubMed
-
- Blevins, J. S., A. F. Gillaspy, T. M. Rechtin, B. K. Hurlburt, and M. S. Smeltzer. 1999. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol. Microbiol. 33:317-326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

