Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus
- PMID: 12218172
- PMCID: PMC129430
- DOI: 10.1073/pnas.182432999
Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus
Abstract
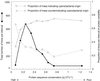
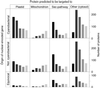
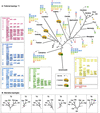
Chloroplasts were once free-living cyanobacteria that became endosymbionts, but the genomes of contemporary plastids encode only approximately 5-10% as many genes as those of their free-living cousins, indicating that many genes were either lost from plastids or transferred to the nucleus during the course of plant evolution. Previous estimates have suggested that between 800 and perhaps as many as 2,000 genes in the Arabidopsis genome might come from cyanobacteria, but genome-wide phylogenetic surveys that could provide direct estimates of this number are lacking. We compared 24,990 proteins encoded in the Arabidopsis genome to the proteins from three cyanobacterial genomes, 16 other prokaryotic reference genomes, and yeast. Of 9,368 Arabidopsis proteins sufficiently conserved for primary sequence comparison, 866 detected homologues only among cyanobacteria and 834 other branched with cyanobacterial homologues in phylogenetic trees. Extrapolating from these conserved proteins to the whole genome, the data suggest that approximately 4,500 of Arabidopsis protein-coding genes ( approximately 18% of the total) were acquired from the cyanobacterial ancestor of plastids. These proteins encompass all functional classes, and the majority of them are targeted to cell compartments other than the chloroplast. Analysis of 15 sequenced chloroplast genomes revealed 117 nuclear-encoded proteins that are also still present in at least one chloroplast genome. A phylogeny of chloroplast genomes inferred from 41 proteins and 8,303 amino acids sites indicates that at least two independent secondary endosymbiotic events have occurred involving red algae and that amino acid composition bias in chloroplast proteins strongly affects plastid genome phylogeny.
Figures
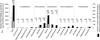
Comment in
-
The genomics of symbiosis: hosts keep the baby and the bath water.Proc Natl Acad Sci U S A. 2002 Sep 17;99(19):11996-7. doi: 10.1073/pnas.202486299. Epub 2002 Sep 9. Proc Natl Acad Sci U S A. 2002. PMID: 12221298 Free PMC article. No abstract available.
References
-
- Goksøyr J. (1967) Nature (London) 214, 1161. - PubMed
-
- Douglas S. E. (1998) Curr. Opin. Gen. Dev. 8, 655-661. - PubMed
-
- Delwiche C. W. (1999) Am. Nat. 154, S164-S177. - PubMed
-
- Tomitani A., Okada, K., Miyashita, H., Matthijs, H. C. P., Ohno, T. & Tanaka, A. (1999) Nature (London) 400, 159-162. - PubMed
-
- Herrmann R. G. (1997) in Eukaryotism and Symbiosis, eds. Schenk, H. E. A., Herrmann, R. G., Jeon, K. W. & Schwemmler, W. (Springer, Heidelberg), pp. 73–118.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

