The hDcp2 protein is a mammalian mRNA decapping enzyme
- PMID: 12218187
- PMCID: PMC130517
- DOI: 10.1073/pnas.192445599
The hDcp2 protein is a mammalian mRNA decapping enzyme
Abstract
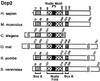
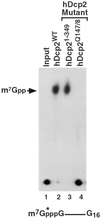
Decapping of mRNA is a critical step in eukaryotic mRNA turnover, yet the proteins involved in this activity remain elusive in mammals. We identified the human Dcp2 protein (hDcp2) as an enzyme containing intrinsic decapping activity. hDcp2 specifically hydrolyzed methylated capped RNA to release m(7)GDP; however, it did not function on the cap structure alone. hDcp2 is therefore functionally distinct from the recently identified mammalian scavenger decapping enzyme, DcpS. hDcp2-mediated decapping required a functional Nudix (nucleotide diphosphate linked to an X moiety) pyrophosphatase motif as mutations in conserved amino acids within this motif disrupted the decapping activity. hDcp2 is detected exclusively in the cytoplasm and predominantly cosediments with polysomes. Consistent with the localization of hDcp2, endogenous Dcp2-like decapping activity was detected in polysomal fractions prepared from mammalian cells. Similar to decapping in yeast, the presence of the poly(A) tail was inhibitory to the endogenous decapping activity, yet unlike yeast, competition of cap-binding proteins by cap analog did not influence the efficiency of decapping. Therefore the mammalian homologue of the yeast Dcp2 protein is an mRNA decapping enzyme demonstrated to contain intrinsic decapping activity.
Figures
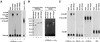
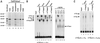
Comment in
-
mRNA decay enzymes: decappers conserved between yeast and mammals.Proc Natl Acad Sci U S A. 2002 Oct 1;99(20):12512-4. doi: 10.1073/pnas.212518099. Epub 2002 Sep 23. Proc Natl Acad Sci U S A. 2002. PMID: 12271148 Free PMC article. No abstract available.
References
-
- Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. Nature (London) 1996;382:642–646. - PubMed
-
- Larimer F W, Hsu C L, Maupin M K, Stevens A. Gene. 1992;120:51–57. - PubMed
-
- Muhlrad D, Decker C J, Parker R. Genes Dev. 1994;8:855–866. - PubMed
-
- Czaplinski K, Ruiz-Echevarria M J, Gonzalez C I, Peltz S W. BioEssays. 1999;21:685–696. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

