Nucleic acid evolution and minimization by nonhomologous random recombination
- PMID: 12219078
- PMCID: PMC2819268
- DOI: 10.1038/nbt736
Nucleic acid evolution and minimization by nonhomologous random recombination
Abstract
We have developed a simple method for exploring nucleic acid sequence space by nonhomologous random recombination (NRR) that enables DNA fragments to randomly recombine in a length-controlled manner without the need for sequence homology. We compared the results of using NRR and error-prone PCR to evolve DNA aptamers that bind streptavidin. Starting with two parental sequences of modest avidin affinity, evolution using NRR resulted in aptamers with 15- to 20-fold higher affinity than the highest-affinity aptamers evolved using error-prone PCR, and 27- or 46-fold higher affinities than parental sequences derived using systematic evolution of ligands by exponential enrichment (SELEX). NRR also facilitates the identification of functional regions within evolved sequences. Inspection of a small number of NRR-evolved clones identified a 40-base DNA sequence, present in multiple copies in each clone, that binds streptavidin. Our findings suggest that NRR may enhance the effectiveness of nucleic acid evolution and the ease of identifying structure-activity relationships among evolved sequences.
Figures



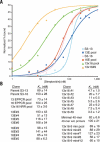


References
-
- Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. - PubMed
-
- Bittker JA, Phillips KJ, Liu DR. Recent advances in the in vitro evolution of nucleic acids. Curr. Opin. Chem. Biol. 2002;6:367–374. - PubMed
-
- Kopylov AM, Spiridonova VA. Combinatorial chemistry of nucleic acids: SELEX. Mol. Biol. 2000;34:940–954. - PubMed
-
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 RNA polymerase. Science. 1990;249:505–510. - PubMed
-
- Cadwell RC, Joyce GF. Mutagenic PCR. PCR Methods Appl. 1994;3:S136–S140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

