The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins
- PMID: 12221072
- PMCID: PMC2173223
- DOI: 10.1083/jcb.200205124
The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins
Abstract
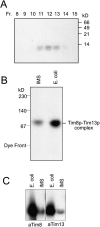
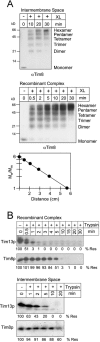
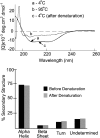
Tim23p is imported via the TIM (translocase of inner membrane)22 pathway for mitochondrial inner membrane proteins. In contrast to precursors with an NH2-terminal targeting presequence that are imported in a linear NH2-terminal manner, we show that Tim23p crosses the outer membrane as a loop before inserting into the inner membrane. The Tim8p-Tim13p complex facilitates translocation across the intermembrane space by binding to the membrane spanning domains as shown by Tim23p peptide scans with the purified Tim8p-Tim13p complex and crosslinking studies with Tim23p fusion constructs. The interaction between Tim23p and the Tim8p-Tim13p complex is not dependent on zinc, and the purified Tim8p-Tim13p complex does not coordinate zinc in the conserved twin CX3C motif. Instead, the cysteine residues seemingly form intramolecular disulfide linkages. Given that proteins of the mitochondrial carrier family also pass through the TOM (translocase of outer membrane) complex as a loop, our study suggests that this translocation mechanism may be conserved. Thus, polytopic inner membrane proteins, which lack an NH2-terminal targeting sequence, pass through the TOM complex as a loop followed by binding of the small Tim proteins to the hydrophobic membrane spanning domains.
Figures




References
-
- Bohni, P.C., G. Daum, and G. Schatz. 1983. Import of proteins into mitochondria. Partial purification of a matrix-located protease involved in cleavage of mitochondrial precursor polypeptides. J. Biol. Chem. 258:4937–4943. - PubMed
-
- Brix, J., S. Rudiger, B. Bukau, J. Schneider-Mergener, and N. Pfanner. 1999. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem. 274:16522–16530. - PubMed
-
- Brix, J., G.A. Ziegler, K. Dietmeier, J. Schneider-Mergener, G.E. Schulz, and N. Pfanner. 2000. The mitochondrial import receptor Tom70: identification of a 25 kDa core domain with a specific binding site for preproteins. J. Mol. Biol. 303:479–488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

