The major sites of cellular phospholipid synthesis and molecular determinants of Fatty Acid and lipid head group specificity
- PMID: 12221122
- PMCID: PMC124149
- DOI: 10.1091/mbc.01-11-0540
The major sites of cellular phospholipid synthesis and molecular determinants of Fatty Acid and lipid head group specificity
Abstract
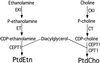
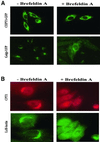
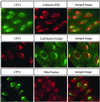
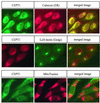
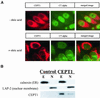
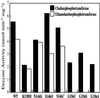
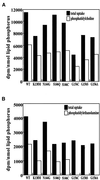
Phosphatidylcholine and phosphatidylethanolamine are the two main phospholipids in eukaryotic cells comprising ~50 and 25% of phospholipid mass, respectively. Phosphatidylcholine is synthesized almost exclusively through the CDP-choline pathway in essentially all mammalian cells. Phosphatidylethanolamine is synthesized through either the CDP-ethanolamine pathway or by the decarboxylation of phosphatidylserine, with the contribution of each pathway being cell type dependent. Two human genes, CEPT1 and CPT1, code for the total compliment of activities that directly synthesize phosphatidylcholine and phosphatidylethanolamine through the CDP-alcohol pathways. CEPT1 transfers a phosphobase from either CDP-choline or CDP-ethanolamine to diacylglycerol to synthesize both phosphatidylcholine and phosphatidylethanolamine, whereas CPT1 synthesizes phosphatidylcholine exclusively. We show through immunofluorescence that brefeldin A treatment relocalizes CPT1, but not CEPT1, implying CPT1 is found in the Golgi. A combination of coimmunofluorescence and subcellular fractionation experiments with various endoplasmic reticulum, Golgi, and nuclear markers confirmed that CPT1 was found in the Golgi and CEPT1 was found in both the endoplasmic reticulum and nuclear membranes. The rate-limiting step for phosphatidylcholine synthesis is catalyzed by the amphitropic CTP:phosphocholine cytidylyltransferase alpha, which is found in the nucleus in most cell types. CTP:phosphocholine cytidylyltransferase alpha is found immediately upstream cholinephosphotransferase, and it translocates from a soluble nuclear location to the nuclear membrane in response to activators of the CDP-choline pathway. Thus, substrate channeling of the CDP-choline produced by CTP:phosphocholine cytidylyltransferase alpha to nuclear located CEPT1 is the mechanism by which upregulation of the CDP-choline pathway increases de novo phosphatidylcholine biosynthesis. In addition, a series of CEPT1 site-directed mutants was generated that allowed for the assignment of specific amino acid residues as structural requirements that directly alter either phospholipid head group or fatty acyl composition. This pinpointed glycine 156 within the catalytic motif as being responsible for the dual CDP-alcohol specificity of CEPT1, whereas mutations within helix 214-228 allowed for the orientation of transmembrane helices surrounding the catalytic site to be definitively positioned.
Figures




References
-
- Ames BN, Dubin DT. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J Biol Chem. 1960;235:769–775. - PubMed
-
- Bae-Lee MS, Carman GM. Phosphatidylserine synthesis in Saccharomyces cerevisiae. Purification and characterization of membrane-associated phosphatidylserine synthase. J Biol Chem. 1984;259:10857–10862. - PubMed
-
- Bankaitis VA, Aitken JF, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

