Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex
- PMID: 12221128
- PMCID: PMC124155
- DOI: 10.1091/mbc.e02-02-0112
Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex
Abstract
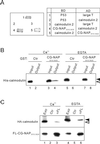
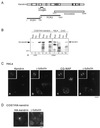
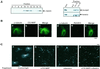
Microtubule assembly is initiated by the gamma-tubulin ring complex (gamma-TuRC). In yeast, the microtubule is nucleated from gamma-TuRC anchored to the amino-terminus of the spindle pole body component Spc110p, which interacts with calmodulin (Cmd1p) at the carboxy-terminus. However, mammalian protein that anchors gamma-TuRC remains to be elucidated. A giant coiled-coil protein, CG-NAP (centrosome and Golgi localized PKN-associated protein), was localized to the centrosome via the carboxyl-terminal region. This region was found to interact with calmodulin by yeast two-hybrid screening, and it shares high homology with the carboxyl-terminal region of another centrosomal coiled-coil protein, kendrin. The amino-terminal region of either CG-NAP or kendrin indirectly associated with gamma-tubulin through binding with gamma-tubulin complex protein 2 (GCP2) and/or GCP3. Furthermore, endogenous CG-NAP and kendrin were coimmunoprecipitated with each other and with endogenous GCP2 and gamma-tubulin, suggesting that CG-NAP and kendrin form complexes and interact with gamma-TuRC in vivo. These proteins were localized to the center of microtubule asters nucleated from isolated centrosomes. Pretreatment of the centrosomes by antibody to CG-NAP or kendrin moderately inhibited the microtubule nucleation; moreover, the combination of these antibodies resulted in stronger inhibition. These results imply that CG-NAP and kendrin provide sites for microtubule nucleation in the mammalian centrosome by anchoring gamma-TuRC.
Figures





Similar articles
-
Centrosome-targeting region of CG-NAP causes centrosome amplification by recruiting cyclin E-cdk2 complex.Genes Cells. 2005 Jan;10(1):75-86. doi: 10.1111/j.1365-2443.2005.00816.x. Genes Cells. 2005. PMID: 15670215
-
Identification of a human centrosomal calmodulin-binding protein that shares homology with pericentrin.Proc Natl Acad Sci U S A. 2000 May 23;97(11):5919-23. doi: 10.1073/pnas.97.11.5919. Proc Natl Acad Sci U S A. 2000. PMID: 10823944 Free PMC article.
-
Characterization of a Drosophila centrosome protein CP309 that shares homology with Kendrin and CG-NAP.Mol Biol Cell. 2004 Jan;15(1):37-45. doi: 10.1091/mbc.e03-03-0191. Epub 2003 Oct 17. Mol Biol Cell. 2004. PMID: 14565985 Free PMC article.
-
Regulation of microtubule nucleation mediated by γ-tubulin complexes.Protoplasma. 2017 May;254(3):1187-1199. doi: 10.1007/s00709-016-1070-z. Epub 2017 Jan 10. Protoplasma. 2017. PMID: 28074286 Review.
-
The structure of the γ-TuRC at the microtubule minus end - not just one solution.Bioessays. 2024 Sep;46(9):e2400117. doi: 10.1002/bies.202400117. Epub 2024 Jul 23. Bioessays. 2024. PMID: 39044599 Review.
Cited by
-
Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation.J Cell Biol. 2006 Feb 13;172(4):517-28. doi: 10.1083/jcb.200511071. J Cell Biol. 2006. PMID: 16476773 Free PMC article.
-
Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly.J Cell Biol. 2004 Aug 30;166(5):637-43. doi: 10.1083/jcb.200405023. J Cell Biol. 2004. PMID: 15337773 Free PMC article.
-
Embryonic expression of pericentrin suggests universal roles in ciliogenesis.Dev Genes Evol. 2006 Sep;216(9):537-42. doi: 10.1007/s00427-006-0065-8. Epub 2006 Mar 14. Dev Genes Evol. 2006. PMID: 16534625
-
Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity.Mol Biol Cell. 2003 Oct;14(10):4260-71. doi: 10.1091/mbc.e02-11-0773. Epub 2003 Jul 11. Mol Biol Cell. 2003. PMID: 14517334 Free PMC article.
-
Moonlighting at the Poles: Non-Canonical Functions of Centrosomes.Front Cell Dev Biol. 2022 Jul 14;10:930355. doi: 10.3389/fcell.2022.930355. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35912107 Free PMC article. Review.
References
-
- Amano M, Mukai H, Ono Y, Chihara K, Matini T, Hamajima Y, Okawa K, Iwanatsu A, Kaibuchi K. Identification of aputative target at Rho as the serine-threonine kinase protein kinase N. Science. 1996;271:648–650. - PubMed
-
- Berchtold MW, Egli R, Rhyner JA, Hameister H, Strehler EE. Localization of the human bona fide calmodulin genes CALM1, CALM2, and CALM3 to chromosomes 14q24–q31, 2p21.1-p21.3, and 19q13.2-q13.3. Genomics. 1993;16:461–465. - PubMed
-
- Diviani D, Langeberg LK, Doxsey SJ, Scott JD. Pericentrin anchors protein kinase A at the centrosome through a newly identified RII-binding domain. Curr Biol. 2000;10:417–420. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

