Gene-expression profile comparisons distinguish seven organs of maize
- PMID: 12225584
- PMCID: PMC126870
- DOI: 10.1186/gb-2002-3-9-research0045
Gene-expression profile comparisons distinguish seven organs of maize
Abstract
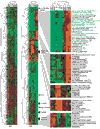
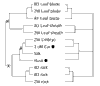
Background: A maize array was fabricated with 5,376 unique expressed sequence tag (EST) clones sequenced from 4-day-old roots, immature ears and adult organ cDNA libraries. To elucidate organ relationships, relative mRNA levels were quantified by hybridization with embryos, three maize vegetative organs (leaf blades, leaf sheaths and roots) from multiple developmental stages, husk leaves and two types of floral organs (immature ears and silks).
Results: Clustering analyses of the hybridization data suggest that maize utilizes both the PEPCK and NADP-ME C(4) photosynthetic routes as genes in these pathways are co-regulated. Husk RNA has a gene-expression profile more similar to floral organs than to vegetative leaves. Only 7% of the genes were highly organ specific, showing over a fourfold difference in at least one of 12 comparisons and 37% showed a two- to fourfold difference. The majority of genes were expressed in diverse organs with little difference in transcript levels. Cross-hybridization among closely related genes within multigene families could obscure tissue specificity. As a first step in elucidating individual gene-expression patterns, we show that 45-nucleotide oligo probes produce signal intensities and signal ratios comparable to PCR probes on the same matrix.
Conclusions: Gene-expression profile studies with cDNA microarrays provide a new molecular tool for defining plant organs and their relationships and for discovering new biological processes in silico. cDNA microarrays are insufficient for differentiating recently duplicated genes. Gene-specific oligo probes printed along with cDNA probes can query individual gene-expression profiles and gene families simultaneously.
Figures





References
-
- Jack T. Plant development going MADS. Plant Mol Biol. 2001;46:515–520. - PubMed
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene-expression patterns with a complementary-DNA microarray. Science. 1995;270:467–470. - PubMed
-
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

