NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria
- PMID: 12225586
- PMCID: PMC126872
- DOI: 10.1186/gb-2002-3-9-research0047
NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria
Abstract
Background: Iron uptake from the host is essential for bacteria that infect animals. To find potential targets for drugs active against pathogenic bacteria, we have searched all completely sequenced genomes of pathogenic bacteria for genes relevant for iron transport.
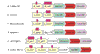
Results: We identified a protein domain that appears in variable copy number in bacterial genes that are usually in the vicinity of a putative Fe3+ siderophore transporter. Accordingly, we have denoted this domain NEAT for 'near transporter'. Most of the bacterial species containing this domain are pathogenic. Sequence features indicate that the domain is anchored to the extracellular side of the membrane. The domain seems to be under high selective pressure for rapid independent duplications that are typical of sequences involved in signaling and binding.
Conclusions: The NEAT domain might be functionally related to iron transport. The taxonomic specificity of this domain and its predicted extracellular position could make it an interesting target for designing new drugs against some highly pathogenic bacteria.
Figures



References
-
- Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. - PubMed
-
- Burkhardt R, Braun V. Nucleotide sequence of the fhuC and fhuD genes involved in iron (III) hydroxamate transport: domains in FhuC homologous to ATP-binding proteins. Mol Gen Genet. 1987;209:49–55. - PubMed
-
- Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

