A new non-linear normalization method for reducing variability in DNA microarray experiments
- PMID: 12225587
- PMCID: PMC126873
- DOI: 10.1186/gb-2002-3-9-research0048
A new non-linear normalization method for reducing variability in DNA microarray experiments
Abstract
Background: Microarray data are subject to multiple sources of variation, of which biological sources are of interest whereas most others are only confounding. Recent work has identified systematic sources of variation that are intensity-dependent and non-linear in nature. Systematic sources of variation are not limited to the differing properties of the cyanine dyes Cy(5) and Cy(3) as observed in cDNA arrays, but are the general case for both oligonucleotide microarray (Affymetrix GeneChips) and cDNA microarray data. Current normalization techniques are most often linear and therefore not capable of fully correcting for these effects.
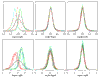
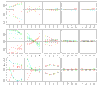
Results: We present here a simple and robust non-linear method for normalization using array signal distribution analysis and cubic splines. These methods compared favorably to normalization using robust local-linear regression (lowess). The application of these methods to oligonucleotide arrays reduced the relative error between replicates by 5-10% compared with a standard global normalization method. Application to cDNA arrays showed improvements over the standard method and over Cy(3)-Cy(5) normalization based on dye-swap replication. In addition, a set of known differentially regulated genes was ranked higher by the t-test. In either cDNA or Affymetrix technology, signal-dependent bias was more than ten times greater than the observed print-tip or spatial effects.
Conclusions: Intensity-dependent normalization is important for both high-density oligonucleotide array and cDNA array data. Both the regression and spline-based methods described here performed better than existing linear methods when assessed on the variability of replicate arrays. Dye-swap normalization was less effective at Cy(3)-Cy(5) normalization than either regression or spline-based methods alone.
Figures










References
-
- Schadt EE, Li C, Su C, Wong WH. Analyzing high-density oligonucleotide gene expression array data. J Cell Biochem. 2000;80:192–202. - PubMed
-
- Schadt EE, Li C, Ellis B, Wong WH. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem. 2001;Suppl 37:120–125. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

