Suprabasin, a novel epidermal differentiation marker and potential cornified envelope precursor
- PMID: 12228223
- PMCID: PMC1283087
- DOI: 10.1074/jbc.M205380200
Suprabasin, a novel epidermal differentiation marker and potential cornified envelope precursor
Abstract
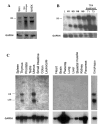
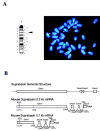
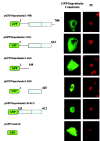
The suprabasin gene is a novel gene expressed in mouse and human differentiating keratinocytes. We identified a partial cDNA encoding suprabasin using a suppression subtractive hybridization method between the proliferative basal and differentiating suprabasal populations of the mouse epidermis. A 3' gene-specific probe hybridized to transcripts of 0.7- and 2.2-kb pairs on Northern blots with specific detection in differentiated keratinocytes of stratified epithelia. The mouse gene was mapped to chromosome 7 by fluorescence in situ hybridization. This region is syntenic to human chromosome band 19q13.1, which contained the only region in the data bases with homology to the mouse suprabasin sequence. During embryonic mouse development, suprabasin mRNA was detected at day 15.5, coinciding with epidermal stratification. Suprabasin was detected in the suprabasal layers of the epithelia in the tongue, stomach, and epidermis. Differentiation of cultured primary epidermal keratinocytes with 0.12 mm Ca(2+) or 12-O-tetradecanoylphorbol-13-acetate treatment resulted in the induction of suprabasin. The 2.2-kb cDNA transcript encodes a protein of 72 kDa with a predicted isoelectric point of 6.85. The translated sequence has an amino-terminal domain, a central domain composed of repeats rich in glycine and alanine, and a carboxyl-terminal domain. The alternatively spliced 0.7-kb transcript encodes a smaller protein that shares the NH(2)- and COOH-terminal regions but lacks the repeat domain region. Cross-linking experiments indicate that suprabasin is a substrate for transglutaminase 2 and 3 activity. Altogether, these results indicate that the suprabasin protein potentially plays a role in the process of epidermal differentiation.
Figures




References
-
- Fuchs E, Byrne C. Curr Opin Genet Dev. 1994;4:725–736. - PubMed
-
- Eckert RL, Welter JF. Mol Biol Rep. 1996;23:59 –70. - PubMed
-
- Steinert PM, Marekov LN, Parry DA. J Biol Chem. 1999;274:1657–1666. - PubMed
-
- Kalinin A, Marekov LN, Steinert PM. J Cell Sci. 2001;114:3069 –3070. - PubMed
-
- Presland RB, Dale BA. Crit Rev Oral Biol Med. 2000;11:383–408. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

