A CaMK cascade activates CRE-mediated transcription in neurons of Caenorhabditis elegans
- PMID: 12231504
- PMCID: PMC1307624
- DOI: 10.1093/embo-reports/kvf191
A CaMK cascade activates CRE-mediated transcription in neurons of Caenorhabditis elegans
Abstract
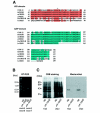
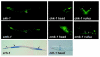
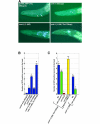
Calcium (Ca2+) signals regulate a diverse set of cellular responses, from proliferation to muscular contraction and neuro-endocrine secretion. The ubiquitous Ca2+ sensor, calmodulin (CaM), translates changes in local intracellular Ca2+ concentrations into changes in enzyme activities. Among its targets, the Ca2+/CaM-dependent protein kinases I and IV (CaMKs) are capable of transducing intraneuronal signals, and these kinases are implicated in neuronal gene regulation that mediates synaptic plasticity in mammals. Recently, the cyclic AMP response element binding protein (CREB) has been proposed as a target for a CaMK cascade involving not only CaMKI or CaMKIV, but also an upstream kinase kinase that is also CaM regulated (CaMKK). Here, we report that all components of this pathway are coexpressed in head neurons of Caenorhabditis elegans. Utilizing a transgenic approach to visualize CREB-dependent transcription in vivo, we show that this CaMK cascade regulates CRE-mediated transcription in a subset of head neurons in living nematodes.
Figures

Similar articles
-
Components of a calmodulin-dependent protein kinase cascade. Molecular cloning, functional characterization and cellular localization of Ca2+/calmodulin-dependent protein kinase kinase beta.J Biol Chem. 1998 Nov 27;273(48):31880-9. doi: 10.1074/jbc.273.48.31880. J Biol Chem. 1998. PMID: 9822657
-
Regulation and function of the calcium/calmodulin-dependent protein kinase IV/protein serine/threonine phosphatase 2A signaling complex.J Biol Chem. 2004 Jul 23;279(30):31708-16. doi: 10.1074/jbc.M404523200. Epub 2004 May 13. J Biol Chem. 2004. PMID: 15143065
-
Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity.Genes Dev. 1994 Nov 1;8(21):2527-39. doi: 10.1101/gad.8.21.2527. Genes Dev. 1994. PMID: 7958915
-
Physiological roles of the Ca2+/CaM-dependent protein kinase cascade in health and disease.Subcell Biochem. 2007;45:169-214. doi: 10.1007/978-1-4020-6191-2_7. Subcell Biochem. 2007. PMID: 18193638 Review.
-
Calmodulin kinases: essential regulators in health and disease.J Neurochem. 2017 Jun;141(6):808-818. doi: 10.1111/jnc.14020. Epub 2017 Apr 17. J Neurochem. 2017. PMID: 28295333 Review.
Cited by
-
The CaM Kinase CMK-1 Mediates a Negative Feedback Mechanism Coupling the C. elegans Glutamate Receptor GLR-1 with Its Own Transcription.PLoS Genet. 2016 Jul 27;12(7):e1006180. doi: 10.1371/journal.pgen.1006180. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27462879 Free PMC article.
-
Regulators of AWC-mediated olfactory plasticity in Caenorhabditis elegans.PLoS Genet. 2009 Dec;5(12):e1000761. doi: 10.1371/journal.pgen.1000761. Epub 2009 Dec 11. PLoS Genet. 2009. PMID: 20011101 Free PMC article.
-
Deep conservation of genes required for both Drosphila melanogaster and Caenorhabditis elegans sleep includes a role for dopaminergic signaling.Sleep. 2014 Sep 1;37(9):1439-51. doi: 10.5665/sleep.3990. Sleep. 2014. PMID: 25142568 Free PMC article.
-
Death Associated Protein Kinase (DAPK) -mediated neurodegenerative mechanisms in nematode excitotoxicity.BMC Neurosci. 2015 Apr 23;16:25. doi: 10.1186/s12868-015-0158-2. BMC Neurosci. 2015. PMID: 25899010 Free PMC article.
-
Interneurons Regulate Locomotion Quiescence via Cyclic Adenosine Monophosphate Signaling During Stress-Induced Sleep in Caenorhabditis elegans.Genetics. 2019 Sep;213(1):267-279. doi: 10.1534/genetics.119.302293. Epub 2019 Jul 10. Genetics. 2019. PMID: 31292211 Free PMC article.
References
-
- Bito H., Deisseroth K. and Tsien R.W. (1996) CREB phosphorylation and dephosphorylation: a Ca2+ and stimulus duration-dependent switch for hippocampal gene expression. Cell, 87, 1203–1214. - PubMed
-
- Eto K., Takahashi N., Kimura Y., Masuho Y., Arai K., Muramatsu M.A. and Tokumitsu H. (1999) Ca2+/calmodulin-dependent protein kinase cascade in Caenorhabditis elegans. Implication in transcriptional activation. J. Biol. Chem., 274, 22556–22562. - PubMed
-
- Gonzalez G.A., Yamamoto K.K., Fischer W.H., Karr D., Menzel P., Biggs W. 3rd, Vale W.W. and Montminy M.R. (1989) A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature, 337, 749–752. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

