Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha
- PMID: 12231507
- PMCID: PMC1307621
- DOI: 10.1093/embo-reports/kvf194
Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha
Abstract
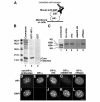
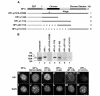
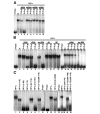
In mammalian cells, as in Schizosaccharomyces pombe and Drosophila, HP1 proteins bind histone H3 tails methylated on lysine 9 (K9). However, whereas K9-methylated H3 histones are distributed throughout the nucleus, HP1 proteins are enriched in pericentromeric heterochromatin. This observation suggests that the methyl-binding property of HP1 may not be sufficient for its heterochromatin targeting. We show that the association of HP1alpha with pericentromeric heterochromatin depends not only on its methyl-binding chromo domain but also on an RNA-binding activity present in the hinge region of the protein that connects the conserved chromo and chromoshadow domains. Our data suggest the existence of complex heterochromatin binding sites composed of methylated histone H3 tails and RNA, with each being recognized by a separate domain of HP1alpha.
Figures


References
-
- Akhmanova A., Verkerk T., Langeveld A., Grosveld F. and Galjart N. (2000) Characterisation of transcriptionally active and inactive chromatin domains in neurons. J. Cell Sci., 113, 4463–4474. - PubMed
-
- Akhtar A., Zink D. and Becker P.B. (2000) Chromodomains are protein–RNA interaction modules. Nature, 407, 405–409. - PubMed
-
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C. and Kouzarides T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. - PubMed
-
- Brodersen D.E., Clemons W.M. Jr, Carter A.P., Wimberly B.T. and Ramakrishnan V. (2002) Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol., 316, 725–768. - PubMed
-
- Eissenberg J.C. and Elgin S.C. (2000) The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev., 10, 204–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

