Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family
- PMID: 12231628
- PMCID: PMC187445
- DOI: 10.1101/gad.1012602
Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family
Abstract
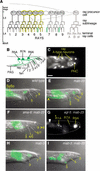
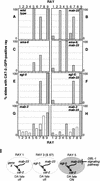
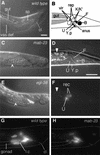
Mutations in Caenorhabditis elegans gene mab-23 cause abnormal male tail morphology and abolish male fecundity but have no obvious effect in the hermaphrodite. Here we show that mab-23 encodes a DM (Doublesex/MAB-3) domain transcription factor necessary for specific aspects of differentiation in sex-specific tissues of the male. mab-23 is required for the patterning of posterior sensory neurons in the male nervous system, sex muscle differentiation, and morphogenesis of the posterior hypodermis, spicules, and proctodeum. Failure of mab-23 mutant males to sire progeny is due primarily to defective sex muscle-mediated turning during copulatory behavior and likely compounded by impairment of sperm passage through the proctodeum. In the male nervous system, mab-23 refines ray neuron subtype distribution by restricting expression of dopaminergic neurotransmitter identity through interactions with the Hox gene egl-5 and a TGF-beta-related signaling pathway. mab-23 has distinct roles and functions independent of mab-3, indicating different aspects of C. elegans male sexual differentiation are coordinated among DM domain family members. Our results support the hypothesis that DM domain genes derive from an ancestral male sexual regulator and suggest how regulation of sexual development has evolved in distinct ways in different phyla.
Figures

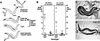
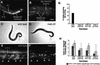

Comment in
-
The remarkable ubiquity of DM domain factors as regulators of sexual phenotype: ancestry or aptitude?Genes Dev. 2002 Sep 15;16(18):2322-6. doi: 10.1101/gad.1025502. Genes Dev. 2002. PMID: 12231620 No abstract available.
Similar articles
-
dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans.Development. 2008 Aug;135(14):2373-82. doi: 10.1242/dev.017046. Epub 2008 Jun 11. Development. 2008. PMID: 18550714 Free PMC article.
-
Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior.Development. 2000 Oct;127(20):4469-80. doi: 10.1242/dev.127.20.4469. Development. 2000. PMID: 11003845
-
The caenorhabditis elegans fate-determining gene mab-9 encodes a T-box protein required to pattern the posterior hindgut.Genes Dev. 2000 Mar 1;14(5):596-603. Genes Dev. 2000. PMID: 10716947 Free PMC article.
-
Somatic sex determination.WormBook. 2006 Feb 10:1-12. doi: 10.1895/wormbook.1.84.1. WormBook. 2006. PMID: 18050479 Free PMC article. Review.
-
RNT-1 regulation in C. elegans.J Cell Biochem. 2005 Sep 1;96(1):8-15. doi: 10.1002/jcb.20423. J Cell Biochem. 2005. PMID: 15988754 Review.
Cited by
-
Initiation of male sperm-transfer behavior in Caenorhabditis elegans requires input from the ventral nerve cord.BMC Biol. 2006 Aug 15;4:26. doi: 10.1186/1741-7007-4-26. BMC Biol. 2006. PMID: 16911797 Free PMC article.
-
Gene Function Prediction Based on Developmental Transcriptomes of the Two Sexes in C. elegans.Cell Rep. 2016 Oct 11;17(3):917-928. doi: 10.1016/j.celrep.2016.09.051. Cell Rep. 2016. PMID: 27732864 Free PMC article.
-
Characterization of the Doublesex/MAB-3 transcription factor DMD-9 in Caenorhabditis elegans.G3 (Bethesda). 2023 Feb 9;13(2):jkac305. doi: 10.1093/g3journal/jkac305. G3 (Bethesda). 2023. PMID: 36454093 Free PMC article.
-
Transcriptome Sequencing Analysis and Functional Identification of Sex Differentiation Genes from the Mosquito Parasitic Nematode, Romanomermis wuchangensis.PLoS One. 2016 Sep 23;11(9):e0163127. doi: 10.1371/journal.pone.0163127. eCollection 2016. PLoS One. 2016. PMID: 27662191 Free PMC article.
-
TGF-β signaling in C. elegans.WormBook. 2013 Jul 10:1-34. doi: 10.1895/wormbook.1.22.2. WormBook. 2013. PMID: 23908056 Free PMC article. Review.
References
-
- Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, control cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. - PubMed
-
- Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. - PubMed
-
- Belote JM, Handler AM, Wolfner MF, Livak KJ, Baker BS. Sex-specific regulation of yolk protein gene expression in Drosophila. Cell. 1985;40:339–348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
