Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans
- PMID: 12231631
- PMCID: PMC187442
- DOI: 10.1101/gad.1011602
Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans
Abstract
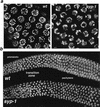
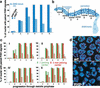
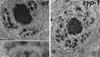
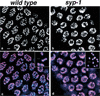
Analysis of Caenorhabditis elegans syp-1 mutants reveals that both synapsis-dependent and -independent mechanisms contribute to stable, productive alignment of homologous chromosomes during meiotic prophase. Early prophase nuclei undergo normal reorganization in syp-1 mutants, and chromosomes initially pair. However, the polarized nuclear organization characteristic of early prophase persists for a prolonged period, and homologs dissociate prematurely; furthermore, the synaptonemal complex (SC) is absent. The predicted structure of SYP-1, its localization at the interface between intimately paired, lengthwise-aligned pachytene homologs, and its kinetics of localization with chromosomes indicate that SYP-1 is an SC structural component. A severe reduction in crossing over together with evidence for accumulated recombination intermediates in syp-1 mutants indicate that initial pairing is not sufficient for completion of exchange and implicates the SC in promoting crossover recombination. Persistence of polarized nuclear organization in syp-1 mutants suggests that SC polymerization may provide a motive force or signal that drives redispersal of chromosomes. Whereas our analysis suggests that the SC is required to stabilize pairing along the entire lengths of chromosomes, striking differences in peak pairing levels for opposite ends of chromosomes in syp-1 mutants reveal the existence of an additional mechanism that can promote local stabilization of pairing, independent of synapsis.
Figures



Similar articles
-
Synaptonemal Complex Central Region Proteins Promote Localization of Pro-crossover Factors to Recombination Events During Caenorhabditis elegans Meiosis.Genetics. 2019 Oct;213(2):395-409. doi: 10.1534/genetics.119.302625. Epub 2019 Aug 20. Genetics. 2019. PMID: 31431470 Free PMC article.
-
SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans.Genetics. 2007 Aug;176(4):2015-25. doi: 10.1534/genetics.107.072413. Epub 2007 Jun 11. Genetics. 2007. PMID: 17565948 Free PMC article.
-
Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis.Genetics. 2007 Aug;176(4):2027-33. doi: 10.1534/genetics.107.076968. Epub 2007 Jun 11. Genetics. 2007. PMID: 17565963 Free PMC article.
-
Targeting Polo-like kinase in space and time during C. elegans meiosis.Cell Cycle. 2021 Aug;20(16):1519-1526. doi: 10.1080/15384101.2021.1953232. Epub 2021 Jul 16. Cell Cycle. 2021. PMID: 34266376 Free PMC article. Review.
-
Meiotic recombination in Caenorhabditis elegans.Chromosome Res. 2007;15(5):607-21. doi: 10.1007/s10577-007-1146-x. Chromosome Res. 2007. PMID: 17674149 Review.
Cited by
-
Full-length synaptonemal complex grows continuously during meiotic prophase in budding yeast.PLoS Genet. 2012;8(10):e1002993. doi: 10.1371/journal.pgen.1002993. Epub 2012 Oct 11. PLoS Genet. 2012. PMID: 23071451 Free PMC article.
-
ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis.Proc Natl Acad Sci U S A. 2021 Apr 6;118(14):e2021671118. doi: 10.1073/pnas.2021671118. Proc Natl Acad Sci U S A. 2021. PMID: 33782125 Free PMC article.
-
Impaired resection of meiotic double-strand breaks channels repair to nonhomologous end joining in Caenorhabditis elegans.Mol Cell Biol. 2013 Jul;33(14):2732-47. doi: 10.1128/MCB.00055-13. Epub 2013 May 13. Mol Cell Biol. 2013. PMID: 23671188 Free PMC article.
-
Synaptonemal Complex Components Are Required for Meiotic Checkpoint Function in Caenorhabditis elegans.Genetics. 2016 Nov;204(3):987-997. doi: 10.1534/genetics.116.191494. Epub 2016 Sep 7. Genetics. 2016. PMID: 27605049 Free PMC article.
-
HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis.Genes Dev. 2005 Nov 15;19(22):2727-43. doi: 10.1101/gad.1338505. Genes Dev. 2005. PMID: 16291646 Free PMC article.
References
-
- Albertson DG, Rose AM, Villeneuve AM. Chromosome organization, mitosis, and meiosis. In: Riddle DL, et al., editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 47–78. - PubMed
-
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
