Proteolytic cleavage of the THR subunit during anaphase limits Drosophila separase function
- PMID: 12231632
- PMCID: PMC187444
- DOI: 10.1101/gad.242202
Proteolytic cleavage of the THR subunit during anaphase limits Drosophila separase function
Abstract
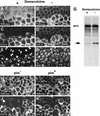
Sister-chromatid separation in mitosis requires proteolytic cleavage of a cohesin subunit. Separase, the corresponding protease, is activated at the metaphase-to-anaphase transition. Activation involves proteolysis of an inhibitory subunit, securin, following ubiquitination mediated by the anaphase-promoting complex/cyclosome. In Drosophila, the securin PIM associates not only with separase (SSE), but also with an additional protein, THR. Here we show that THR is cleaved after the metaphase-to-anaphase transition. THR cleavage only occurs in functional SSE complexes and in a region that matches the separase cleavage-site consensus. Mutations in this region abolish mitotic THR cleavage. These results indicate that THR is cleaved by SSE. Expression of noncleavable THR variants results in cold-sensitive maternal-effect lethality. This lethality can be suppressed by a reduction of catalytically active SSE levels, indicating that THR cleavage inactivates SSE complexes. THR cleavage is particularly important during the process of cellularization, which follows completion of the last syncytial mitosis of early embryogenesis, suggesting that Drosophila separase has other targets in addition to cohesin subunits.
Figures




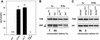
Similar articles
-
Drosophila separase is required for sister chromatid separation and binds to PIM and THR.Genes Dev. 2001 Oct 1;15(19):2572-84. doi: 10.1101/gad.207301. Genes Dev. 2001. PMID: 11581162 Free PMC article.
-
Genetic interactions between Cdk1-CyclinB and the Separase complex in Drosophila.Development. 2005 Apr;132(8):1875-84. doi: 10.1242/dev.01780. Epub 2005 Mar 16. Development. 2005. PMID: 15772129
-
Drosophila securin destruction involves a D-box and a KEN-box and promotes anaphase in parallel with Cyclin A degradation.J Cell Sci. 2003 Jun 15;116(Pt 12):2453-60. doi: 10.1242/jcs.00411. Epub 2003 Apr 30. J Cell Sci. 2003. PMID: 12724352
-
[Activity of separase and its regulation].Yi Chuan. 2004 May;26(3):383-6. Yi Chuan. 2004. PMID: 15640025 Review. Chinese.
-
How do so few control so many?Cell. 2005 Mar 25;120(6):739-46. doi: 10.1016/j.cell.2005.03.006. Cell. 2005. PMID: 15797376 Review.
Cited by
-
Chromosome separation during Drosophila male meiosis I requires separase-mediated cleavage of the homolog conjunction protein UNO.PLoS Genet. 2020 Oct 1;16(10):e1008928. doi: 10.1371/journal.pgen.1008928. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33001976 Free PMC article.
-
How noncrossover homologs are conjoined and segregated in Drosophila male meiosis I: Stable but reversible homolog linkers require a novel Separase target protein.PLoS Genet. 2020 Oct 1;16(10):e1008997. doi: 10.1371/journal.pgen.1008997. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33002007 Free PMC article. No abstract available.
-
What is your assay for sister-chromatid cohesion?EMBO J. 2007 Nov 14;26(22):4609-18. doi: 10.1038/sj.emboj.7601898. Epub 2007 Oct 25. EMBO J. 2007. PMID: 17962808 Free PMC article. Review. No abstract available.
-
PP2A delays APC/C-dependent degradation of separase-associated but not free securin.EMBO J. 2014 May 16;33(10):1134-47. doi: 10.1002/embj.201488098. Epub 2014 Apr 29. EMBO J. 2014. PMID: 24781523 Free PMC article.
-
Live Cell Monitoring of Separase Activity, a Key Enzymatic Reaction for Chromosome Segregation, with Chimeric FRET-Based Molecular Sensor upon Cell Cycle Progression.Biosensors (Basel). 2024 Apr 15;14(4):192. doi: 10.3390/bios14040192. Biosensors (Basel). 2024. PMID: 38667185 Free PMC article.
References
-
- Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. - PubMed
-
- Brinkley BR, Cartwright J., Jr Cold-labile and cold-stable microtubules in the mitotic spindle of mammalian cells. Ann NY Acad Sci. 1975;253:428–439. - PubMed
-
- Chen L, Puri R, Lefkowitz EJ, Kakar SS. Identification of the human pituitary tumor transforming gene (hPTTG) family: Molecular structure, expression, and chromosomal localization. Gene. 2000;248:41–50. - PubMed
-
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. - PubMed
-
- Clarkson M, Saint R. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 1999;18:457–462. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
