RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase
- PMID: 12234935
- PMCID: PMC126294
- DOI: 10.1093/emboj/cdf498
RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase
Abstract
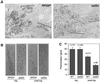
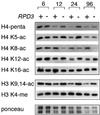
rRNA transcription in Saccharomyces cerevisiae is performed by RNA polymerase I and regulated by changes in growth conditions. During log phase, approximately 50% of the ribosomal DNA (rDNA) genes in each cell are transcribed and maintained in an open, psoralen-accessible conformation. During stationary phase, the percentage of open rDNA genes is greatly reduced. In this study we found that the Rpd3 histone deacetylase was required to inactivate (close) individual rDNA genes as cells entered stationary phase. Even though approximately 50% of the rDNA genes remained open during stationary phase in rpd3Delta mutants, overall rRNA synthesis was still reduced. Using electron microscopy of Miller chromatin spreads, we found that the number of RNA polymerases transcribing each open gene in the rpd3Delta mutant was significantly reduced when cells grew past log phase. Bulk levels of histone H3 and H4 acetylation were reduced during stationary phase in an RPD3-dependent manner. However, histone H3 and H4 acetylation was not significantly altered at the rDNA locus in an rpd3Delta mutant. Rpd3 therefore regulates the number of open rDNA repeats.
Figures



References
-
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (2000) Current Protocols in Molecular Biology, Vol. 2. John Wiley & Sons, New York, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

