Interaction of HCF-1 with a cellular nuclear export factor
- PMID: 12235138
- PMCID: PMC4291127
- DOI: 10.1074/jbc.M205440200
Interaction of HCF-1 with a cellular nuclear export factor
Abstract
HCF-1 is a cellular protein required by VP16 to activate the herpes simplex virus (HSV) immediate-early genes. VP16 is a component of the viral tegument and, after release into the cell, binds to HCF-1 and translocates to the nucleus to form a complex with the POU domain protein Oct-1 and a VP16-responsive DNA sequence. This VP16-induced complex boosts transcription of the viral immediate-early genes and initiates lytic replication. In uninfected cells, HCF-1 functions as a coactivator for the cellular transcription factors LZIP and GABP and also plays an essential role in cell proliferation. VP16 and LZIP share a tetrapeptide HCF-binding motif recognized by the beta-propeller domain of HCF-1. Here we describe a new cellular HCF-1 beta-propeller domain binding protein, termed HPIP, which contains a functional HCF-binding motif and a leucine-rich nuclear export sequence. We show that HPIP shuttles between the nucleus and cytoplasm in a CRM1-dependent manner and that overexpression of HPIP leads to accumulation of HCF-1 in the cytoplasm. These data suggest that HPIP regulates HCF-1 activity by modulating its subcellular localization. Furthermore, HPIP-mediated export may provide the pool of cytoplasmic HCF-1 required for import of virion-derived VP16 into the nucleus.
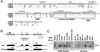
Figures






References
-
- Roizman B, Sears AE. In: Fields Virology. 3rd Ed. Fields BN, Knipe DM, Howley PM, editors. Vol. 2. Philadelphia, PA: Lippincott-Raven Publishers; 1996. pp. 2231–2295.
-
- O’Hare P. Semin. Virol. 1993;4:145–155.
-
- Wilson AC, Cleary MA, Lai J-S, LaMarco K, Peterson MG, Herr W. Cold Spring Harbor Symp. Quant. Biol. 1993;58:167–178. - PubMed
-
- Thompson CC, McKnight SL. Trends Genet. 1992;8:232–236.
-
- Stern S, Herr W. Genes Dev. 1991;5:2555–2566. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

