Interaction of the ocr gene 0.3 protein of bacteriophage T7 with EcoKI restriction/modification enzyme
- PMID: 12235377
- PMCID: PMC137103
- DOI: 10.1093/nar/gkf518
Interaction of the ocr gene 0.3 protein of bacteriophage T7 with EcoKI restriction/modification enzyme
Abstract
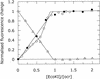
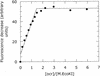
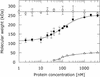
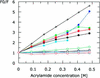
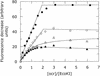
The ocr protein, the product of gene 0.3 of bacteriophage T7, is a structural mimic of the phosphate backbone of B-form DNA. In total it mimics 22 phosphate groups over approximately 24 bp of DNA. This mimicry allows it to block DNA binding by type I DNA restriction enzymes and to inhibit these enzymes. We have determined that multiple ocr dimers can bind stoichiometrically to the archetypal type I enzyme, EcoKI. One dimer binds to the core methyltransferase and two to the complete bifunctional restriction and modification enzyme. Ocr can also bind to the component subunits of EcoKI. Binding affinity to the methyltransferase core is extremely strong with a large favourable enthalpy change and an unfavourable entropy change. This strong interaction prevents the dissociation of the methyltransferase which occurs upon dilution of the enzyme. This stabilisation arises because the interaction appears to involve virtually the entire surface area of ocr and leads to the enzyme completely wrapping around ocr.
Figures
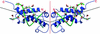




References
-
- Walkinshaw M.D., Taylor,P., Sturrock,S.S., Atanasiu,C., Berge,T., Henderson,R.M., Edwardson,J.M. and Dryden,D.T.F. (2002) Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell, 9, 187–194. - PubMed
-
- Zavilgelsky G.B. (2000) Antirestriction. Mol. Biol. (Mosk.), 34, 854–862. - PubMed
-
- Studier F.W. (1975) Gene 0.3 of bacteriophage T7 acts to overcome the DNA restriction system of the host. J. Mol. Biol., 94, 283–295. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

